Adrian Galdran
AI for Mycetoma Diagnosis in Histopathological Images: The MICCAI 2024 Challenge
Dec 25, 2025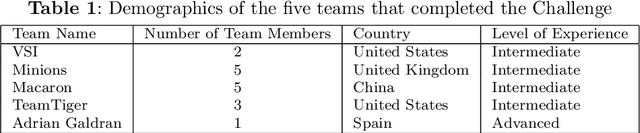
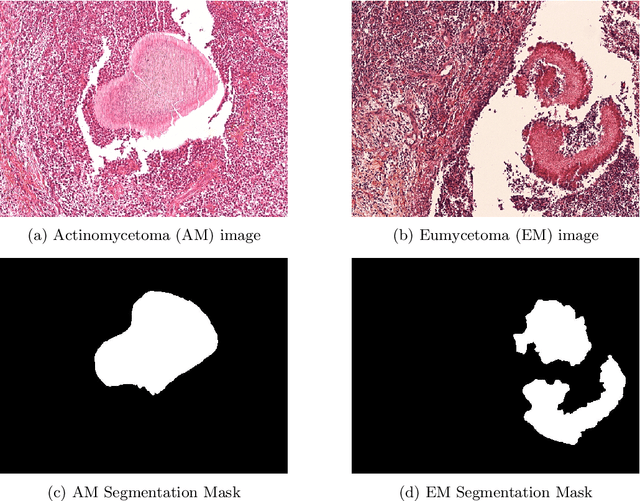
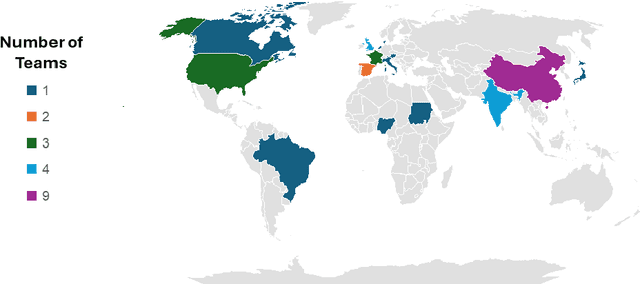
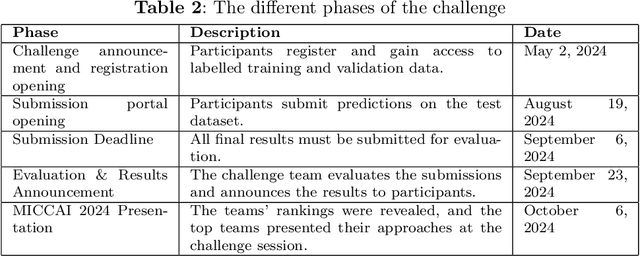
Abstract:Mycetoma is a neglected tropical disease caused by fungi or bacteria leading to severe tissue damage and disabilities. It affects poor and rural communities and presents medical challenges and socioeconomic burdens on patients and healthcare systems in endemic regions worldwide. Mycetoma diagnosis is a major challenge in mycetoma management, particularly in low-resource settings where expert pathologists are limited. To address this challenge, this paper presents an overview of the Mycetoma MicroImage: Detect and Classify Challenge (mAIcetoma) which was organized to advance mycetoma diagnosis through AI solutions. mAIcetoma focused on developing automated models for segmenting mycetoma grains and classifying mycetoma types from histopathological images. The challenge attracted the attention of several teams worldwide to participate and five finalist teams fulfilled the challenge objectives. The teams proposed various deep learning architectures for the ultimate goal of this challenge. Mycetoma database (MyData) was provided to participants as a standardized dataset to run the proposed models. Those models were evaluated using evaluation metrics. Results showed that all the models achieved high segmentation accuracy, emphasizing the necessitate of grain detection as a critical step in mycetoma diagnosis. In addition, the top-performing models show a significant performance in classifying mycetoma types.
Calibration and Uncertainty for multiRater Volume Assessment in multiorgan Segmentation (CURVAS) challenge results
May 13, 2025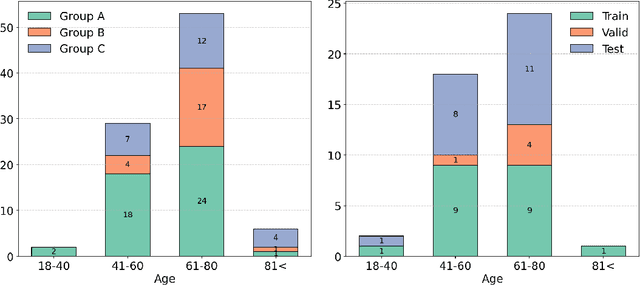
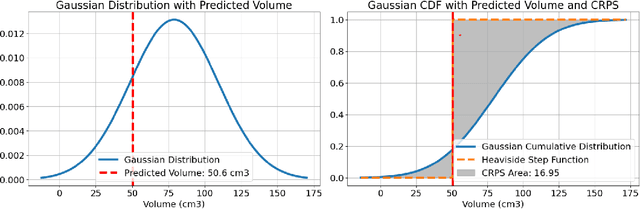

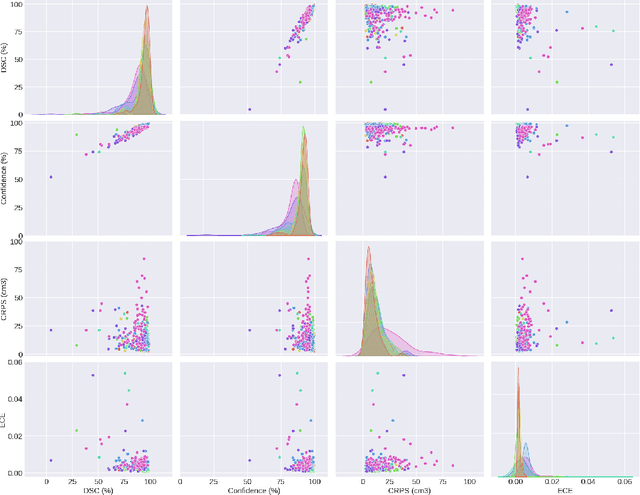
Abstract:Deep learning (DL) has become the dominant approach for medical image segmentation, yet ensuring the reliability and clinical applicability of these models requires addressing key challenges such as annotation variability, calibration, and uncertainty estimation. This is why we created the Calibration and Uncertainty for multiRater Volume Assessment in multiorgan Segmentation (CURVAS), which highlights the critical role of multiple annotators in establishing a more comprehensive ground truth, emphasizing that segmentation is inherently subjective and that leveraging inter-annotator variability is essential for robust model evaluation. Seven teams participated in the challenge, submitting a variety of DL models evaluated using metrics such as Dice Similarity Coefficient (DSC), Expected Calibration Error (ECE), and Continuous Ranked Probability Score (CRPS). By incorporating consensus and dissensus ground truth, we assess how DL models handle uncertainty and whether their confidence estimates align with true segmentation performance. Our findings reinforce the importance of well-calibrated models, as better calibration is strongly correlated with the quality of the results. Furthermore, we demonstrate that segmentation models trained on diverse datasets and enriched with pre-trained knowledge exhibit greater robustness, particularly in cases deviating from standard anatomical structures. Notably, the best-performing models achieved high DSC and well-calibrated uncertainty estimates. This work underscores the need for multi-annotator ground truth, thorough calibration assessments, and uncertainty-aware evaluations to develop trustworthy and clinically reliable DL-based medical image segmentation models.
KPIs 2024 Challenge: Advancing Glomerular Segmentation from Patch- to Slide-Level
Feb 11, 2025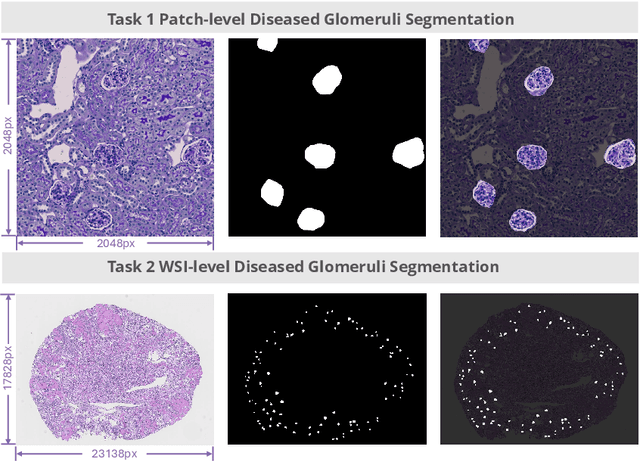
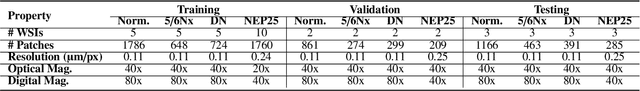
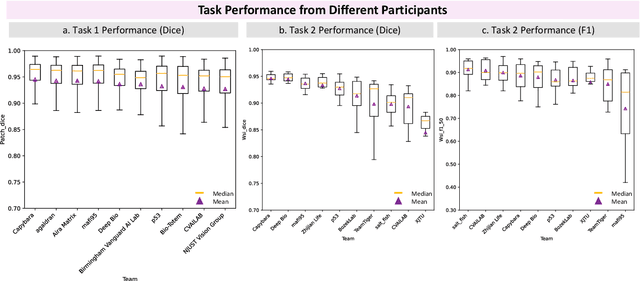

Abstract:Chronic kidney disease (CKD) is a major global health issue, affecting over 10% of the population and causing significant mortality. While kidney biopsy remains the gold standard for CKD diagnosis and treatment, the lack of comprehensive benchmarks for kidney pathology segmentation hinders progress in the field. To address this, we organized the Kidney Pathology Image Segmentation (KPIs) Challenge, introducing a dataset that incorporates preclinical rodent models of CKD with over 10,000 annotated glomeruli from 60+ Periodic Acid Schiff (PAS)-stained whole slide images. The challenge includes two tasks, patch-level segmentation and whole slide image segmentation and detection, evaluated using the Dice Similarity Coefficient (DSC) and F1-score. By encouraging innovative segmentation methods that adapt to diverse CKD models and tissue conditions, the KPIs Challenge aims to advance kidney pathology analysis, establish new benchmarks, and enable precise, large-scale quantification for disease research and diagnosis.
Data-Centric Label Smoothing for Explainable Glaucoma Screening from Eye Fundus Images
Jun 06, 2024Abstract:As current computing capabilities increase, modern machine learning and computer vision system tend to increase in complexity, mostly by means of larger models and advanced optimization strategies. Although often neglected, in many problems there is also much to be gained by considering potential improvements in understanding and better leveraging already-available training data, including annotations. This so-called data-centric approach can lead to substantial performance increases, sometimes beyond what can be achieved by larger models. In this paper we adopt such an approach for the task of justifiable glaucoma screening from retinal images. In particular, we focus on how to combine information from multiple annotators of different skills into a tailored label smoothing scheme that allows us to better employ a large collection of fundus images, instead of discarding samples suffering from inter-rater variability. Internal validation results indicate that our bespoke label smoothing approach surpasses the performance of a standard resnet50 model and also the same model trained with conventional label smoothing techniques, in particular for the multi-label scenario of predicting clinical reasons of glaucoma likelihood in a highly imbalanced screening context. Our code is made available at github.com/agaldran/justraigs .
Polyp and Surgical Instrument Segmentation with Double Encoder-Decoder Networks
Jun 06, 2024

Abstract:This paper describes a solution for the MedAI competition, in which participants were required to segment both polyps and surgical instruments from endoscopic images. Our approach relies on a double encoder-decoder neural network which we have previously applied for polyp segmentation, but with a series of enhancements: a more powerful encoder architecture, an improved optimization procedure, and the post-processing of segmentations based on tempered model ensembling. Experimental results show that our method produces segmentations that show a good agreement with manual delineations provided by medical experts.
Benchmarking the CoW with the TopCoW Challenge: Topology-Aware Anatomical Segmentation of the Circle of Willis for CTA and MRA
Dec 29, 2023



Abstract:The Circle of Willis (CoW) is an important network of arteries connecting major circulations of the brain. Its vascular architecture is believed to affect the risk, severity, and clinical outcome of serious neuro-vascular diseases. However, characterizing the highly variable CoW anatomy is still a manual and time-consuming expert task. The CoW is usually imaged by two angiographic imaging modalities, magnetic resonance angiography (MRA) and computed tomography angiography (CTA), but there exist limited public datasets with annotations on CoW anatomy, especially for CTA. Therefore we organized the TopCoW Challenge in 2023 with the release of an annotated CoW dataset and invited submissions worldwide for the CoW segmentation task, which attracted over 140 registered participants from four continents. TopCoW dataset was the first public dataset with voxel-level annotations for CoW's 13 vessel components, made possible by virtual-reality (VR) technology. It was also the first dataset with paired MRA and CTA from the same patients. TopCoW challenge aimed to tackle the CoW characterization problem as a multiclass anatomical segmentation task with an emphasis on topological metrics. The top performing teams managed to segment many CoW components to Dice scores around 90%, but with lower scores for communicating arteries and rare variants. There were also topological mistakes for predictions with high Dice scores. Additional topological analysis revealed further areas for improvement in detecting certain CoW components and matching CoW variant's topology accurately. TopCoW represented a first attempt at benchmarking the CoW anatomical segmentation task for MRA and CTA, both morphologically and topologically.
Performance Metrics for Probabilistic Ordinal Classifiers
Sep 15, 2023



Abstract:Ordinal classification models assign higher penalties to predictions further away from the true class. As a result, they are appropriate for relevant diagnostic tasks like disease progression prediction or medical image grading. The consensus for assessing their categorical predictions dictates the use of distance-sensitive metrics like the Quadratic-Weighted Kappa score or the Expected Cost. However, there has been little discussion regarding how to measure performance of probabilistic predictions for ordinal classifiers. In conventional classification, common measures for probabilistic predictions are Proper Scoring Rules (PSR) like the Brier score, or Calibration Errors like the ECE, yet these are not optimal choices for ordinal classification. A PSR named Ranked Probability Score (RPS), widely popular in the forecasting field, is more suitable for this task, but it has received no attention in the image analysis community. This paper advocates the use of the RPS for image grading tasks. In addition, we demonstrate a counter-intuitive and questionable behavior of this score, and propose a simple fix for it. Comprehensive experiments on four large-scale biomedical image grading problems over three different datasets show that the RPS is a more suitable performance metric for probabilistic ordinal predictions. Code to reproduce our experiments can be found at https://github.com/agaldran/prob_ord_metrics .
Why is the winner the best?
Mar 30, 2023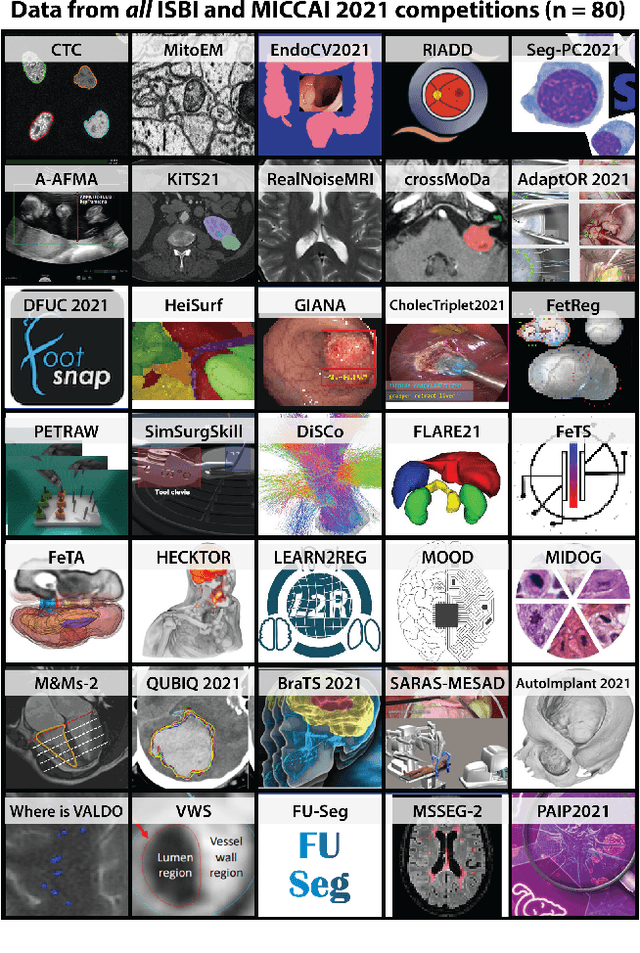
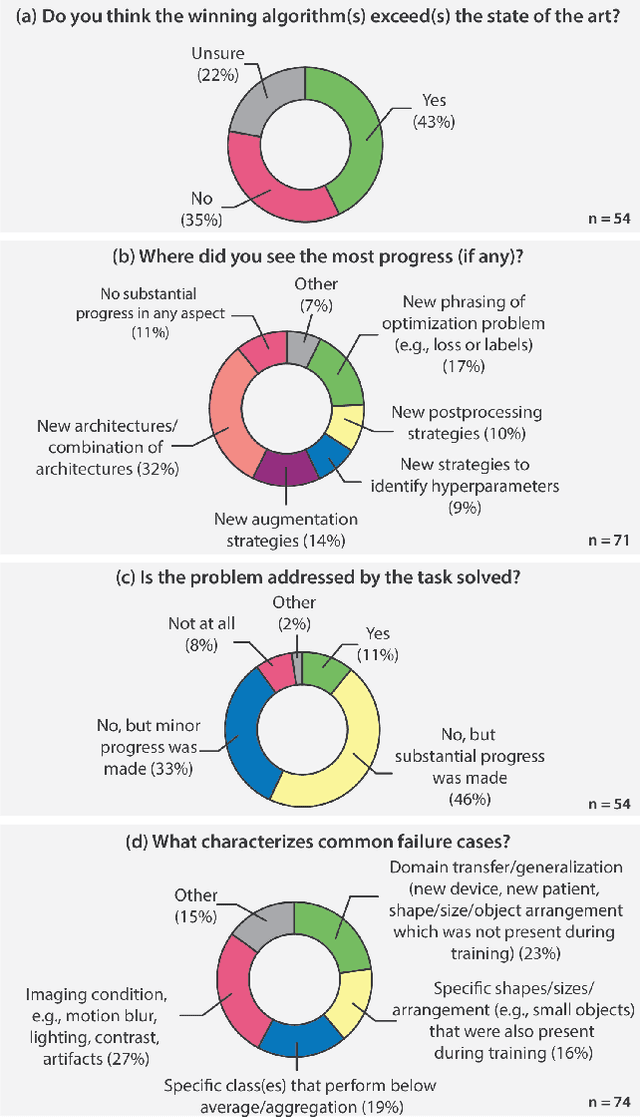
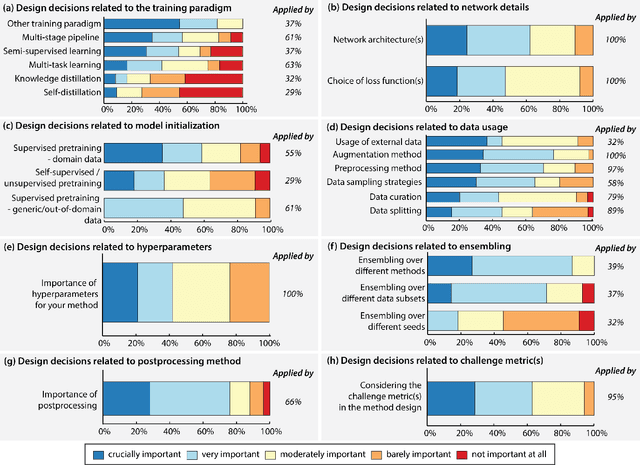
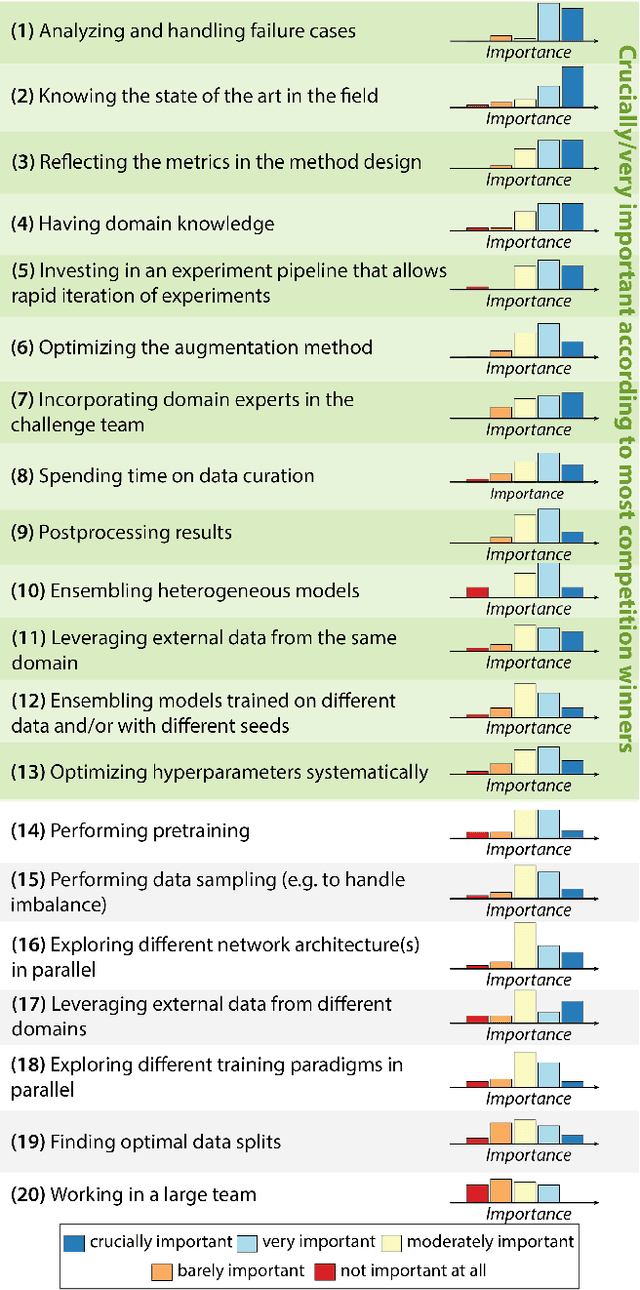
Abstract:International benchmarking competitions have become fundamental for the comparative performance assessment of image analysis methods. However, little attention has been given to investigating what can be learnt from these competitions. Do they really generate scientific progress? What are common and successful participation strategies? What makes a solution superior to a competing method? To address this gap in the literature, we performed a multi-center study with all 80 competitions that were conducted in the scope of IEEE ISBI 2021 and MICCAI 2021. Statistical analyses performed based on comprehensive descriptions of the submitted algorithms linked to their rank as well as the underlying participation strategies revealed common characteristics of winning solutions. These typically include the use of multi-task learning (63%) and/or multi-stage pipelines (61%), and a focus on augmentation (100%), image preprocessing (97%), data curation (79%), and postprocessing (66%). The "typical" lead of a winning team is a computer scientist with a doctoral degree, five years of experience in biomedical image analysis, and four years of experience in deep learning. Two core general development strategies stood out for highly-ranked teams: the reflection of the metrics in the method design and the focus on analyzing and handling failure cases. According to the organizers, 43% of the winning algorithms exceeded the state of the art but only 11% completely solved the respective domain problem. The insights of our study could help researchers (1) improve algorithm development strategies when approaching new problems, and (2) focus on open research questions revealed by this work.
Multi-Head Multi-Loss Model Calibration
Mar 02, 2023Abstract:Delivering meaningful uncertainty estimates is essential for a successful deployment of machine learning models in the clinical practice. A central aspect of uncertainty quantification is the ability of a model to return predictions that are well-aligned with the actual probability of the model being correct, also known as model calibration. Although many methods have been proposed to improve calibration, no technique can match the simple, but expensive approach of training an ensemble of deep neural networks. In this paper we introduce a form of simplified ensembling that bypasses the costly training and inference of deep ensembles, yet it keeps its calibration capabilities. The idea is to replace the common linear classifier at the end of a network by a set of heads that are supervised with different loss functions to enforce diversity on their predictions. Specifically, each head is trained to minimize a weighted Cross-Entropy loss, but the weights are different among the different branches. We show that the resulting averaged predictions can achieve excellent calibration without sacrificing accuracy in two challenging datasets for histopathological and endoscopic image classification. Our experiments indicate that Multi-Head Multi-Loss classifiers are inherently well-calibrated, outperforming other recent calibration techniques and even challenging Deep Ensembles' performance. Code to reproduce our experiments can be found at \url{https://github.com/agaldran/mhml_calibration} .
AIROGS: Artificial Intelligence for RObust Glaucoma Screening Challenge
Feb 10, 2023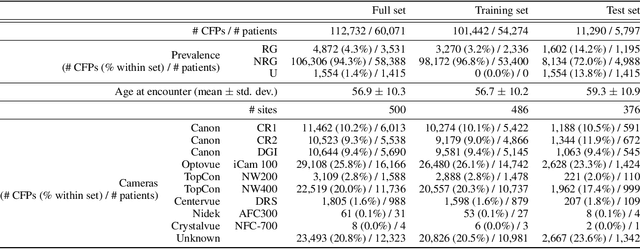
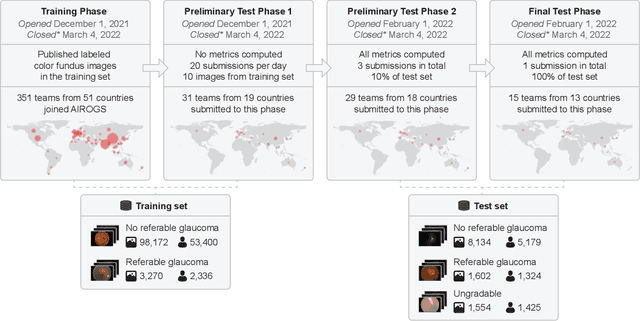
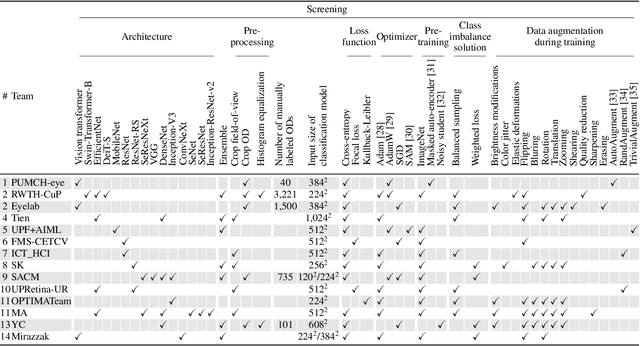
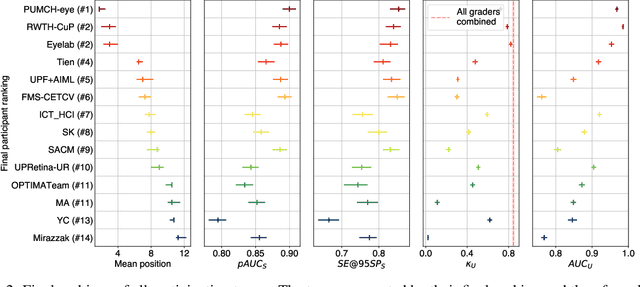
Abstract:The early detection of glaucoma is essential in preventing visual impairment. Artificial intelligence (AI) can be used to analyze color fundus photographs (CFPs) in a cost-effective manner, making glaucoma screening more accessible. While AI models for glaucoma screening from CFPs have shown promising results in laboratory settings, their performance decreases significantly in real-world scenarios due to the presence of out-of-distribution and low-quality images. To address this issue, we propose the Artificial Intelligence for Robust Glaucoma Screening (AIROGS) challenge. This challenge includes a large dataset of around 113,000 images from about 60,000 patients and 500 different screening centers, and encourages the development of algorithms that are robust to ungradable and unexpected input data. We evaluated solutions from 14 teams in this paper, and found that the best teams performed similarly to a set of 20 expert ophthalmologists and optometrists. The highest-scoring team achieved an area under the receiver operating characteristic curve of 0.99 (95% CI: 0.98-0.99) for detecting ungradable images on-the-fly. Additionally, many of the algorithms showed robust performance when tested on three other publicly available datasets. These results demonstrate the feasibility of robust AI-enabled glaucoma screening.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge