Quan Huu Cap
From Brainwaves to Brain Scans: A Robust Neural Network for EEG-to-fMRI Synthesis
Feb 11, 2025Abstract:While functional magnetic resonance imaging (fMRI) offers rich spatial resolution, it is limited by high operational costs and significant infrastructural demands. In contrast, electroencephalography (EEG) provides millisecond-level precision in capturing electrical activity but lacks the spatial resolution necessary for precise neural localization. To bridge these gaps, we introduce E2fNet, a simple yet effective deep learning model for synthesizing fMRI images from low-cost EEG data. E2fNet is specifically designed to capture and translate meaningful features from EEG across electrode channels into accurate fMRI representations. Extensive evaluations across three datasets demonstrate that E2fNet consistently outperforms existing methods, achieving state-of-the-art results in terms of the structural similarity index measure (SSIM). Our findings suggest that E2fNet is a promising, cost-effective solution for enhancing neuroimaging capabilities. The code is available at https://github.com/kgr20/E2fNet.
KPIs 2024 Challenge: Advancing Glomerular Segmentation from Patch- to Slide-Level
Feb 11, 2025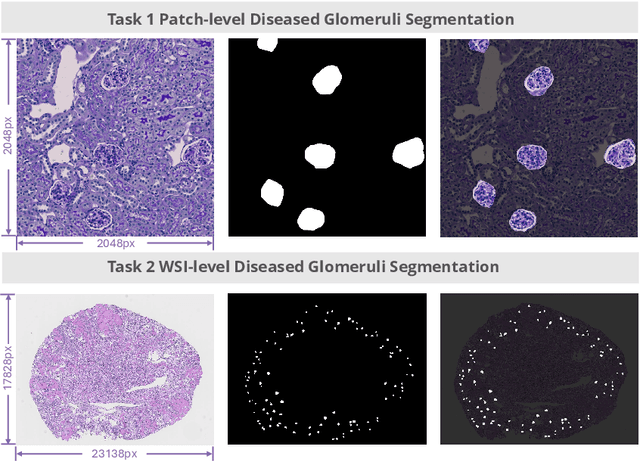
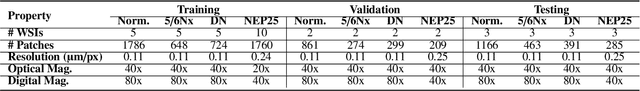
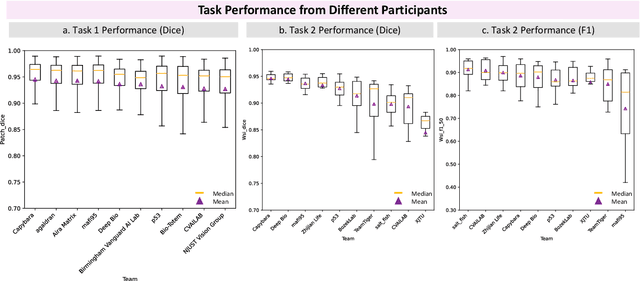

Abstract:Chronic kidney disease (CKD) is a major global health issue, affecting over 10% of the population and causing significant mortality. While kidney biopsy remains the gold standard for CKD diagnosis and treatment, the lack of comprehensive benchmarks for kidney pathology segmentation hinders progress in the field. To address this, we organized the Kidney Pathology Image Segmentation (KPIs) Challenge, introducing a dataset that incorporates preclinical rodent models of CKD with over 10,000 annotated glomeruli from 60+ Periodic Acid Schiff (PAS)-stained whole slide images. The challenge includes two tasks, patch-level segmentation and whole slide image segmentation and detection, evaluated using the Dice Similarity Coefficient (DSC) and F1-score. By encouraging innovative segmentation methods that adapt to diverse CKD models and tissue conditions, the KPIs Challenge aims to advance kidney pathology analysis, establish new benchmarks, and enable precise, large-scale quantification for disease research and diagnosis.
Color-Quality Invariance for Robust Medical Image Segmentation
Feb 11, 2025Abstract:Single-source domain generalization (SDG) in medical image segmentation remains a significant challenge, particularly for images with varying color distributions and qualities. Previous approaches often struggle when models trained on high-quality images fail to generalize to low-quality test images due to these color and quality shifts. In this work, we propose two novel techniques to enhance generalization: dynamic color image normalization (DCIN) module and color-quality generalization (CQG) loss. The DCIN dynamically normalizes the color of test images using two reference image selection strategies. Specifically, the DCIN utilizes a global reference image selection (GRIS), which finds a universal reference image, and a local reference image selection (LRIS), which selects a semantically similar reference image per test sample. Additionally, CQG loss enforces invariance to color and quality variations by ensuring consistent segmentation predictions across transformed image pairs. Experimental results show that our proposals significantly improve segmentation performance over the baseline on two target domain datasets, despite being trained solely on a single source domain. Notably, our model achieved up to a 32.3-point increase in Dice score compared to the baseline, consistently producing robust and usable results even under substantial domain shifts. Our work contributes to the development of more robust medical image segmentation models that generalize across unseen domains. The implementation code is available at https://github.com/RaviShah1/DCIN-CQG.
An Effective Pipeline for Whole-Slide Image Glomerulus Segmentation
Nov 07, 2024Abstract:Whole-slide images (WSI) glomerulus segmentation is essential for accurately diagnosing kidney diseases. In this work, we propose a practical pipeline for glomerulus segmentation that effectively enhances both patch-level and WSI-level segmentation tasks. Our approach leverages stitching on overlapping patches, increasing the detection coverage, especially when glomeruli are located near patch image borders. In addition, we conduct comprehensive evaluations from different segmentation models across two large and diverse datasets with over 30K glomerulus annotations. Experimental results demonstrate that models using our pipeline outperform the previous state-of-the-art method, achieving superior results across both datasets and setting a new benchmark for glomerulus segmentation in WSIs. The code and pre-trained models are available at https://github.com/huuquan1994/wsi_glomerulus_seg.
High-Quality Medical Image Generation from Free-hand Sketch
Feb 01, 2024Abstract:Generating medical images from human-drawn free-hand sketches holds promise for various important medical imaging applications. Due to the extreme difficulty in collecting free-hand sketch data in the medical domain, most deep learning-based methods have been proposed to generate medical images from the synthesized sketches (e.g., edge maps or contours of segmentation masks from real images). However, these models often fail to generalize on the free-hand sketches, leading to unsatisfactory results. In this paper, we propose a practical free-hand sketch-to-image generation model called Sketch2MedI that learns to represent sketches in StyleGAN's latent space and generate medical images from it. Thanks to the ability to encode sketches into this meaningful representation space, Sketch2MedI only requires synthesized sketches for training, enabling a cost-effective learning process. Our Sketch2MedI demonstrates a robust generalization to free-hand sketches, resulting in high-quality and realistic medical image generations. Comparative evaluations of Sketch2MedI against the pix2pix, CycleGAN, UNIT, and U-GAT-IT models show superior performance in generating pharyngeal images, both quantitative and qualitative across various metrics.
Towards Robust Plant Disease Diagnosis with Hard-sample Re-mining Strategy
Sep 05, 2023Abstract:With rich annotation information, object detection-based automated plant disease diagnosis systems (e.g., YOLO-based systems) often provide advantages over classification-based systems (e.g., EfficientNet-based), such as the ability to detect disease locations and superior classification performance. One drawback of these detection systems is dealing with unannotated healthy data with no real symptoms present. In practice, healthy plant data appear to be very similar to many disease data. Thus, those models often produce mis-detected boxes on healthy images. In addition, labeling new data for detection models is typically time-consuming. Hard-sample mining (HSM) is a common technique for re-training a model by using the mis-detected boxes as new training samples. However, blindly selecting an arbitrary amount of hard-sample for re-training will result in the degradation of diagnostic performance for other diseases due to the high similarity between disease and healthy data. In this paper, we propose a simple but effective training strategy called hard-sample re-mining (HSReM), which is designed to enhance the diagnostic performance of healthy data and simultaneously improve the performance of disease data by strategically selecting hard-sample training images at an appropriate level. Experiments based on two practical in-field eight-class cucumber and ten-class tomato datasets (42.7K and 35.6K images) show that our HSReM training strategy leads to a substantial improvement in the overall diagnostic performance on large-scale unseen data. Specifically, the object detection model trained using the HSReM strategy not only achieved superior results as compared to the classification-based state-of-the-art EfficientNetV2-Large model and the original object detection model, but also outperformed the model using the HSM strategy.
A Practical Framework for Unsupervised Structure Preservation Medical Image Enhancement
Apr 04, 2023Abstract:Medical images are extremely valuable for supporting medical diagnoses. However, in practice, low-quality (LQ) medical images, such as images that are hazy/blurry, have uneven illumination, or are out of focus, among others, are often obtained during data acquisition. This leads to difficulties in the screening and diagnosis of medical diseases. Several generative adversarial networks (GAN)-based image enhancement methods have been proposed and have shown promising results. However, there is a quality-originality trade-off among these methods in the sense that they produce visually pleasing results but lose the ability to preserve originality, especially the structural inputs. Moreover, to our knowledge, there is no objective metric in evaluating the structure preservation of medical image enhancement methods in unsupervised settings due to the unavailability of paired ground-truth data. In this study, we propose a framework for practical unsupervised medical image enhancement that includes (1) a non-reference objective evaluation of structure preservation for medical image enhancement tasks called Laplacian structural similarity index measure (LaSSIM), which is based on SSIM and the Laplacian pyramid, and (2) a novel unsupervised GAN-based method called Laplacian medical image enhancement (LaMEGAN) to support the improvement of both originality and quality from LQ images. The LaSSIM metric does not require clean reference images and has been shown to be superior to SSIM in capturing image structural changes under image degradations, such as strong blurring on different datasets. The experiments demonstrated that our LaMEGAN achieves a satisfactory balance between quality and originality, with robust structure preservation performance while generating compelling visual results with very high image quality scores. The code will be made available at https://github.com/AillisInc/USPMIE.
Bulk Production Augmentation Towards Explainable Melanoma Diagnosis
Mar 03, 2021



Abstract:Although highly accurate automated diagnostic techniques for melanoma have been reported, the realization of a system capable of providing diagnostic evidence based on medical indices remains an open issue because of difficulties in obtaining reliable training data. In this paper, we propose bulk production augmentation (BPA) to generate high-quality, diverse pseudo-skin tumor images with the desired structural malignant features for additional training images from a limited number of labeled images. The proposed BPA acts as an effective data augmentation in constructing the feature detector for the atypical pigment network (APN), which is a key structure in melanoma diagnosis. Experiments show that training with images generated by our BPA largely boosts the APN detection performance by 20.0 percentage points in the area under the receiver operating characteristic curve, which is 11.5 to 13.7 points higher than that of conventional CycleGAN-based augmentations in AUC.
MIINet: An Image Quality Improvement Framework for Supporting Medical Diagnosis
Nov 28, 2020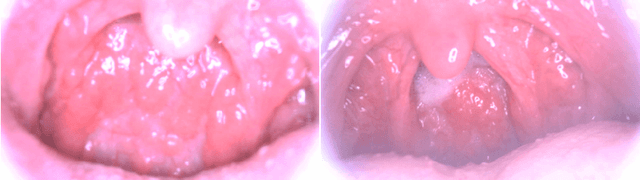
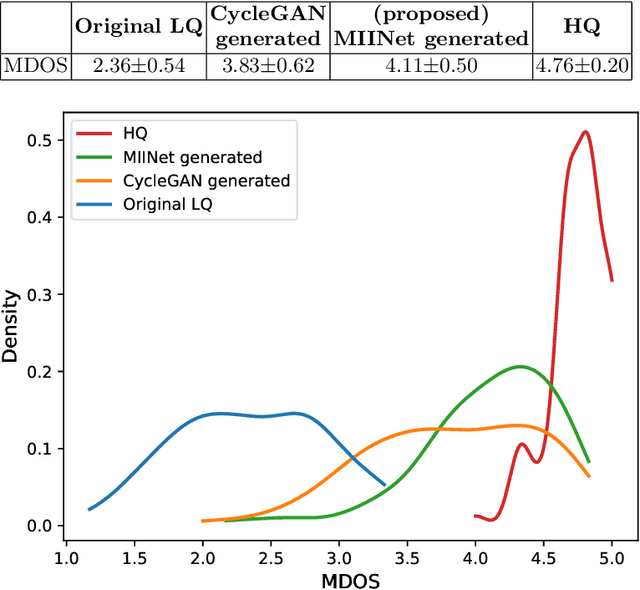
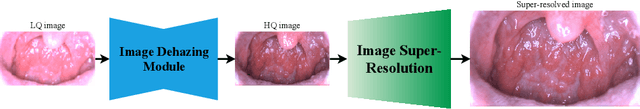
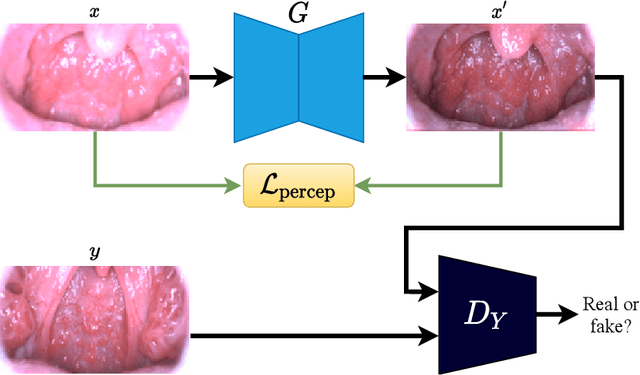
Abstract:Medical images have been indispensable and useful tools for supporting medical experts in making diagnostic decisions. However, taken medical images especially throat and endoscopy images are normally hazy, lack of focus, or uneven illumination. Thus, these could difficult the diagnosis process for doctors. In this paper, we propose MIINet, a novel image-to-image translation network for improving quality of medical images by unsupervised translating low-quality images to the high-quality clean version. Our MIINet is not only capable of generating high-resolution clean images, but also preserving the attributes of original images, making the diagnostic more favorable for doctors. Experiments on dehazing 100 practical throat images show that our MIINet largely improves the mean doctor opinion score (MDOS), which assesses the quality and the reproducibility of the images from the baseline of 2.36 to 4.11, while dehazed images by CycleGAN got lower score of 3.83. The MIINet is confirmed by three physicians to be satisfying in supporting throat disease diagnostic from original low-quality images.
LASSR: Effective Super-Resolution Method for Plant Disease Diagnosis
Oct 12, 2020Abstract:The collection of high-resolution training data is crucial in building robust plant disease diagnosis systems, since such data have a significant impact on diagnostic performance. However, they are very difficult to obtain and are not always available in practice. Deep learning-based techniques, and particularly generative adversarial networks (GANs), can be applied to generate high-quality super-resolution images, but these methods often produce unexpected artifacts that can lower the diagnostic performance. In this paper, we propose a novel artifact-suppression super-resolution method that is specifically designed for diagnosing leaf disease, called Leaf Artifact-Suppression Super Resolution (LASSR). Thanks to its own artifact removal module that detects and suppresses artifacts to a considerable extent, LASSR can generate much more pleasing, high-quality images compared to the state-of-the-art ESRGAN model. Experiments based on a five-class cucumber disease (including healthy) discrimination model show that training with data generated by LASSR significantly boosts the performance on an unseen test dataset by nearly 22% compared with the baseline, and that our approach is more than 2% better than a model trained with images generated by ESRGAN.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge