Satoshi Kagiwada
DDD: Discriminative Difficulty Distance for plant disease diagnosis
Jan 01, 2025



Abstract:Recent studies on plant disease diagnosis using machine learning (ML) have highlighted concerns about the overestimated diagnostic performance due to inappropriate data partitioning, where training and test datasets are derived from the same source (domain). Plant disease diagnosis presents a challenging classification task, characterized by its fine-grained nature, vague symptoms, and the extensive variability of image features within each domain. In this study, we propose the concept of Discriminative Difficulty Distance (DDD), a novel metric designed to quantify the domain gap between training and test datasets while assessing the classification difficulty of test data. DDD provides a valuable tool for identifying insufficient diversity in training data, thus supporting the development of more diverse and robust datasets. We investigated multiple image encoders trained on different datasets and examined whether the distances between datasets, measured using low-dimensional representations generated by the encoders, are suitable as a DDD metric. The study utilized 244,063 plant disease images spanning four crops and 34 disease classes collected from 27 domains. As a result, we demonstrated that even if the test images are from different crops or diseases than those used to train the encoder, incorporating them allows the construction of a distance measure for a dataset that strongly correlates with the difficulty of diagnosis indicated by the disease classifier developed independently. Compared to the base encoder, pre-trained only on ImageNet21K, the correlation higher by 0.106 to 0.485, reaching a maximum of 0.909.
Few-shot Metric Domain Adaptation: Practical Learning Strategies for an Automated Plant Disease Diagnosis
Dec 25, 2024



Abstract:Numerous studies have explored image-based automated systems for plant disease diagnosis, demonstrating impressive diagnostic capabilities. However, recent large-scale analyses have revealed a critical limitation: that the diagnostic capability suffers significantly when validated on images captured in environments (domains) differing from those used during training. This shortfall stems from the inherently limited dataset size and the diverse manifestation of disease symptoms, combined with substantial variations in cultivation environments and imaging conditions, such as equipment and composition. These factors lead to insufficient variety in training data, ultimately constraining the system's robustness and generalization. To address these challenges, we propose Few-shot Metric Domain Adaptation (FMDA), a flexible and effective approach for enhancing diagnostic accuracy in practical systems, even when only limited target data is available. FMDA reduces domain discrepancies by introducing a constraint to the diagnostic model that minimizes the "distance" between feature spaces of source (training) data and target data with limited samples. FMDA is computationally efficient, requiring only basic feature distance calculations and backpropagation, and can be seamlessly integrated into any machine learning (ML) pipeline. In large-scale experiments, involving 223,015 leaf images across 20 fields and 3 crop species, FMDA achieved F1 score improvements of 11.1 to 29.3 points compared to cases without target data, using only 10 images per disease from the target domain. Moreover, FMDA consistently outperformed fine-tuning methods utilizing the same data, with an average improvement of 8.5 points.
Hierarchical Object Detection and Recognition Framework for Practical Plant Disease Diagnosis
Jul 25, 2024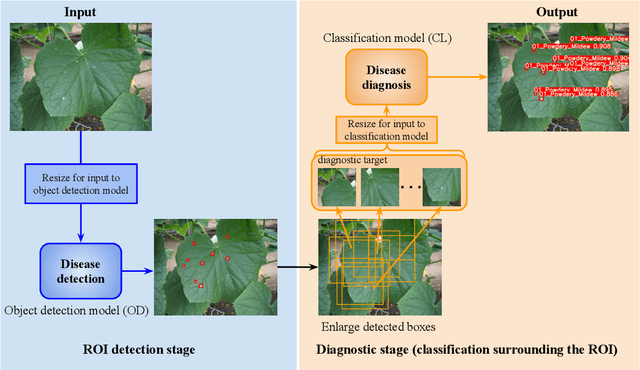
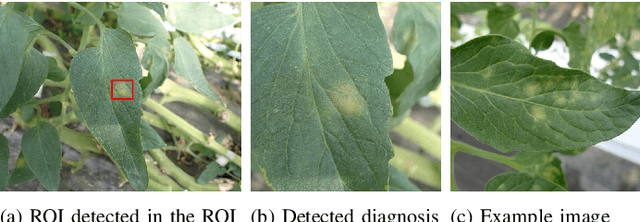

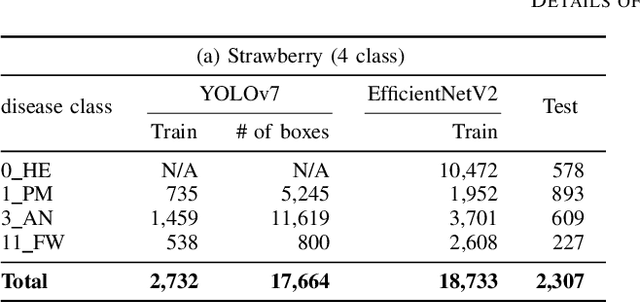
Abstract:Recently, object detection methods (OD; e.g., YOLO-based models) have been widely utilized in plant disease diagnosis. These methods demonstrate robustness to distance variations and excel at detecting small lesions compared to classification methods (CL; e.g., CNN models). However, there are issues such as low diagnostic performance for hard-to-detect diseases and high labeling costs. Additionally, since healthy cases cannot be explicitly trained, there is a risk of false positives. We propose the Hierarchical object detection and recognition framework (HODRF), a sophisticated and highly integrated two-stage system that combines the strengths of both OD and CL for plant disease diagnosis. In the first stage, HODRF uses OD to identify regions of interest (ROIs) without specifying the disease. In the second stage, CL diagnoses diseases surrounding the ROIs. HODRF offers several advantages: (1) Since OD detects only one type of ROI, HODRF can detect diseases with limited training images by leveraging its ability to identify other lesions. (2) While OD over-detects healthy cases, HODRF significantly reduces these errors by using CL in the second stage. (3) CL's accuracy improves in HODRF as it identifies diagnostic targets given as ROIs, making it less vulnerable to size changes. (4) HODRF benefits from CL's lower annotation costs, allowing it to learn from a larger number of images. We implemented HODRF using YOLOv7 for OD and EfficientNetV2 for CL and evaluated its performance on a large-scale dataset (4 crops, 20 diseased and healthy classes, 281K images). HODRF outperformed YOLOv7 alone by 5.8 to 21.5 points on healthy data and 0.6 to 7.5 points on macro F1 scores, and it improved macro F1 by 1.1 to 7.2 points over EfficientNetV2.
Investigation to answer three key questions concerning plant pest identification and development of a practical identification framework
Jul 25, 2024Abstract:The development of practical and robust automated diagnostic systems for identifying plant pests is crucial for efficient agricultural production. In this paper, we first investigate three key research questions (RQs) that have not been addressed thus far in the field of image-based plant pest identification. Based on the knowledge gained, we then develop an accurate, robust, and fast plant pest identification framework using 334K images comprising 78 combinations of four plant portions (the leaf front, leaf back, fruit, and flower of cucumber, tomato, strawberry, and eggplant) and 20 pest species captured at 27 farms. The results reveal the following. (1) For an appropriate evaluation of the model, the test data should not include images of the field from which the training images were collected, or other considerations to increase the diversity of the test set should be taken into account. (2) Pre-extraction of ROIs, such as leaves and fruits, helps to improve identification accuracy. (3) Integration of closely related species using the same control methods and cross-crop training methods for the same pests, are effective. Our two-stage plant pest identification framework, enabling ROI detection and convolutional neural network (CNN)-based identification, achieved a highly practical performance of 91.0% and 88.5% in mean accuracy and macro F1 score, respectively, for 12,223 instances of test data of 21 classes collected from unseen fields, where 25 classes of images from 318,971 samples were used for training; the average identification time was 476 ms/image.
* 40 pages, 10 figures
Towards Robust Plant Disease Diagnosis with Hard-sample Re-mining Strategy
Sep 05, 2023Abstract:With rich annotation information, object detection-based automated plant disease diagnosis systems (e.g., YOLO-based systems) often provide advantages over classification-based systems (e.g., EfficientNet-based), such as the ability to detect disease locations and superior classification performance. One drawback of these detection systems is dealing with unannotated healthy data with no real symptoms present. In practice, healthy plant data appear to be very similar to many disease data. Thus, those models often produce mis-detected boxes on healthy images. In addition, labeling new data for detection models is typically time-consuming. Hard-sample mining (HSM) is a common technique for re-training a model by using the mis-detected boxes as new training samples. However, blindly selecting an arbitrary amount of hard-sample for re-training will result in the degradation of diagnostic performance for other diseases due to the high similarity between disease and healthy data. In this paper, we propose a simple but effective training strategy called hard-sample re-mining (HSReM), which is designed to enhance the diagnostic performance of healthy data and simultaneously improve the performance of disease data by strategically selecting hard-sample training images at an appropriate level. Experiments based on two practical in-field eight-class cucumber and ten-class tomato datasets (42.7K and 35.6K images) show that our HSReM training strategy leads to a substantial improvement in the overall diagnostic performance on large-scale unseen data. Specifically, the object detection model trained using the HSReM strategy not only achieved superior results as compared to the classification-based state-of-the-art EfficientNetV2-Large model and the original object detection model, but also outperformed the model using the HSM strategy.
LASSR: Effective Super-Resolution Method for Plant Disease Diagnosis
Oct 12, 2020Abstract:The collection of high-resolution training data is crucial in building robust plant disease diagnosis systems, since such data have a significant impact on diagnostic performance. However, they are very difficult to obtain and are not always available in practice. Deep learning-based techniques, and particularly generative adversarial networks (GANs), can be applied to generate high-quality super-resolution images, but these methods often produce unexpected artifacts that can lower the diagnostic performance. In this paper, we propose a novel artifact-suppression super-resolution method that is specifically designed for diagnosing leaf disease, called Leaf Artifact-Suppression Super Resolution (LASSR). Thanks to its own artifact removal module that detects and suppresses artifacts to a considerable extent, LASSR can generate much more pleasing, high-quality images compared to the state-of-the-art ESRGAN model. Experiments based on a five-class cucumber disease (including healthy) discrimination model show that training with data generated by LASSR significantly boosts the performance on an unseen test dataset by nearly 22% compared with the baseline, and that our approach is more than 2% better than a model trained with images generated by ESRGAN.
LeafGAN: An Effective Data Augmentation Method for Practical Plant Disease Diagnosis
Feb 24, 2020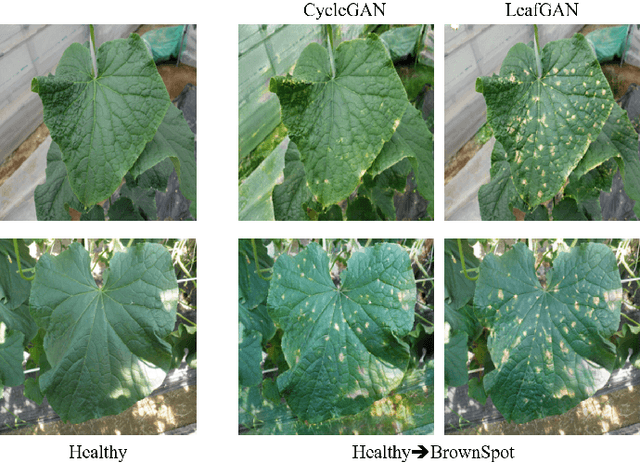
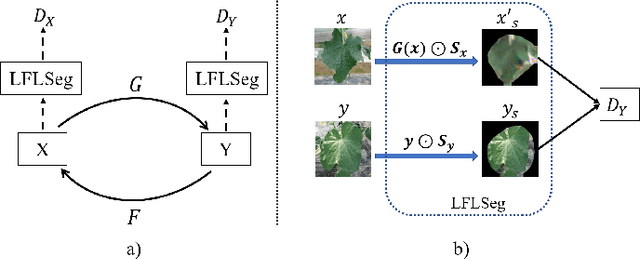

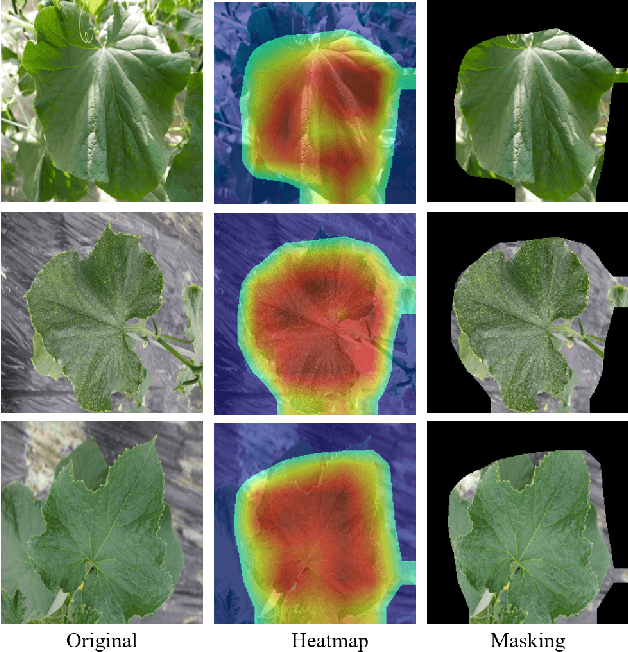
Abstract:Many applications for the automated diagnosis of plant disease have been developed based on the success of deep learning techniques. However, these applications often suffer from overfitting, and the diagnostic performance is drastically decreased when used on test datasets from new environments. The typical reasons for this are that the symptoms to be detected are unclear or faint, and there are limitations related to data diversity. In this paper, we propose LeafGAN, a novel image-to-image translation system with own attention mechanism. LeafGAN generates a wide variety of diseased images via transformation from healthy images, as a data augmentation tool for improving the performance of plant disease diagnosis. Thanks to its own attention mechanism, our model can transform only relevant areas from images with a variety of backgrounds, thus enriching the versatility of the training images. Experiments with five-class cucumber disease classification show that data augmentation with vanilla CycleGAN cannot help to improve the generalization, i.e. disease diagnostic performance increased by only 0.7% from the baseline. In contrast, LeafGAN boosted the diagnostic performance by 7.4%. We also visually confirmed the generated images by our LeafGAN were much better quality and more convincing than those generated by vanilla CycleGAN.
Super-Resolution for Practical Automated Plant Disease Diagnosis System
Nov 26, 2019
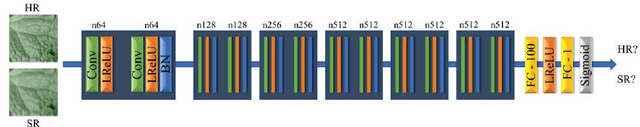


Abstract:Automated plant diagnosis using images taken from a distance is often insufficient in resolution and degrades diagnostic accuracy since the important external characteristics of symptoms are lost. In this paper, we first propose an effective pre-processing method for improving the performance of automated plant disease diagnosis systems using super-resolution techniques. We investigate the efficiency of two different super-resolution methods by comparing the disease diagnostic performance on the practical original high-resolution, low-resolution, and super-resolved cucumber images. Our method generates super-resolved images that look very close to natural images with 4$\times$ upscaling factors and is capable of recovering the lost detailed symptoms, largely boosting the diagnostic performance. Our model improves the disease classification accuracy by 26.9% over the bicubic interpolation method of 65.6% and shows a small gap (3% lower) between the original result of 95.5%.
AOP: An Anti-overfitting Pretreatment for Practical Image-based Plant Diagnosis
Nov 25, 2019

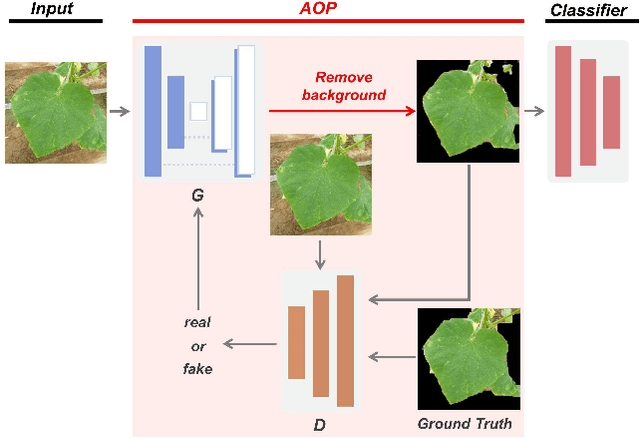
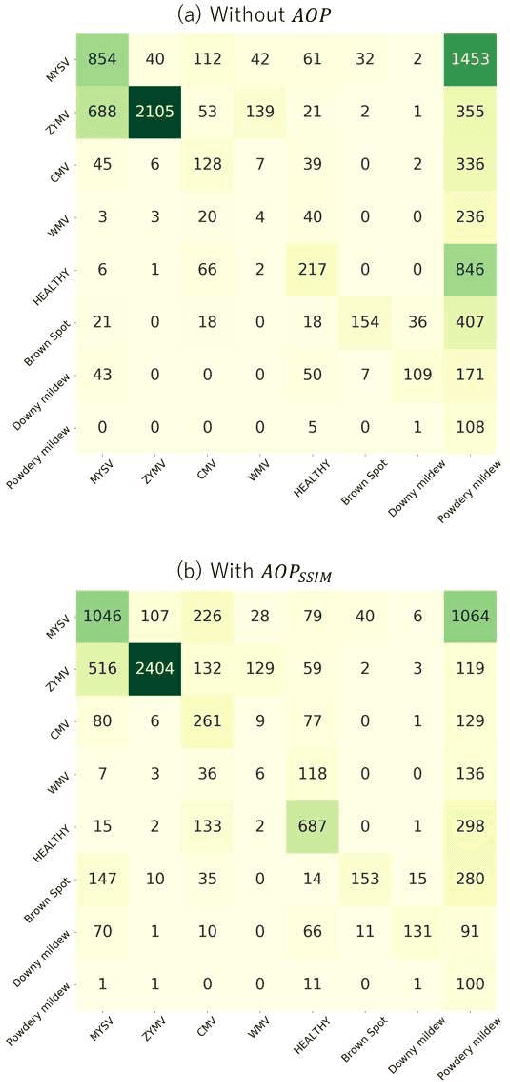
Abstract:In image-based plant diagnosis, clues related to diagnosis are often unclear, and the other factors such as image backgrounds often have a significant impact on the final decision. As a result, overfitting due to latent similarities in the dataset often occurs, and the diagnostic performance on real unseen data (e,g. images from other farms) is usually dropped significantly. However, this problem has not been sufficiently explored, since many systems have shown excellent diagnostic performance due to the bias caused by the similarities in the dataset. In this study, we investigate this problem with experiments using more than 50,000 images of cucumber leaves, and propose an anti-overfitting pretreatment (AOP) for realizing practical image-based plant diagnosis systems. The AOP detects the area of interest (leaf, fruit etc.) and performs brightness calibration as a preprocessing step. The experimental results demonstrate that our AOP can improve the accuracy of diagnosis for unknown test images from different farms by 12.2% in a practical setting.
A comparable study: Intrinsic difficulties of practical plant diagnosis from wide-angle images
Nov 22, 2019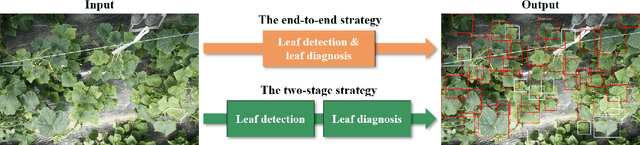


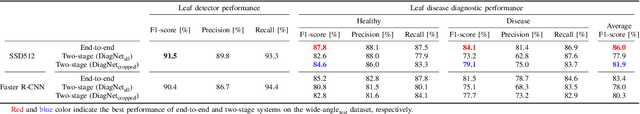
Abstract:Practical automated detection and diagnosis of plant disease from wide-angle images (i.e. in-field images containing multiple leaves using a fixed-position camera) is a very important application for large-scale farm management, in view of the need to ensure global food security. However, developing automated systems for disease diagnosis is often difficult, because labeling a reliable wide-angle disease dataset from actual field images is very laborious. In addition, the potential similarities between the training and test data lead to a serious problem of model overfitting. In this paper, we investigate changes in performance when applying disease diagnosis systems to different scenarios involving wide-angle cucumber test data captured on real farms, and propose an effective diagnostic strategy. We show that leading object recognition techniques such as SSD and Faster R-CNN achieve excellent end-to-end disease diagnostic performance only for a test dataset that is collected from the same population as the training dataset (with F1-score of 81.5% - 84.1% for diagnosed cases of disease), but their performance markedly deteriorates for a completely different test dataset (with F1-score of 4.4 - 6.2%). In contrast, our proposed two-stage systems using independent leaf detection and leaf diagnosis stages attain a promising disease diagnostic performance that is more than six times higher than end-to-end systems (with F1-score of 33.4 - 38.9%) on an unseen target dataset. We also confirm the efficiency of our proposal based on visual assessment, concluding that a two-stage model is a suitable and reasonable choice for practical applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge