Hiroki Tani
LASSR: Effective Super-Resolution Method for Plant Disease Diagnosis
Oct 12, 2020Abstract:The collection of high-resolution training data is crucial in building robust plant disease diagnosis systems, since such data have a significant impact on diagnostic performance. However, they are very difficult to obtain and are not always available in practice. Deep learning-based techniques, and particularly generative adversarial networks (GANs), can be applied to generate high-quality super-resolution images, but these methods often produce unexpected artifacts that can lower the diagnostic performance. In this paper, we propose a novel artifact-suppression super-resolution method that is specifically designed for diagnosing leaf disease, called Leaf Artifact-Suppression Super Resolution (LASSR). Thanks to its own artifact removal module that detects and suppresses artifacts to a considerable extent, LASSR can generate much more pleasing, high-quality images compared to the state-of-the-art ESRGAN model. Experiments based on a five-class cucumber disease (including healthy) discrimination model show that training with data generated by LASSR significantly boosts the performance on an unseen test dataset by nearly 22% compared with the baseline, and that our approach is more than 2% better than a model trained with images generated by ESRGAN.
Super-Resolution for Practical Automated Plant Disease Diagnosis System
Nov 26, 2019
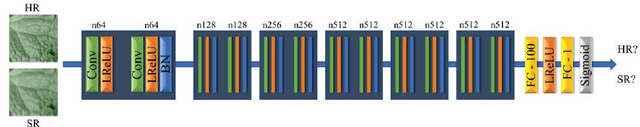


Abstract:Automated plant diagnosis using images taken from a distance is often insufficient in resolution and degrades diagnostic accuracy since the important external characteristics of symptoms are lost. In this paper, we first propose an effective pre-processing method for improving the performance of automated plant disease diagnosis systems using super-resolution techniques. We investigate the efficiency of two different super-resolution methods by comparing the disease diagnostic performance on the practical original high-resolution, low-resolution, and super-resolved cucumber images. Our method generates super-resolved images that look very close to natural images with 4$\times$ upscaling factors and is capable of recovering the lost detailed symptoms, largely boosting the diagnostic performance. Our model improves the disease classification accuracy by 26.9% over the bicubic interpolation method of 65.6% and shows a small gap (3% lower) between the original result of 95.5%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge