Hitoshi Iyatomi
Acquisition of interpretable domain information during brain MR image harmonization for content-based image retrieval
Oct 16, 2025Abstract:Medical images like MR scans often show domain shifts across imaging sites due to scanner and protocol differences, which degrade machine learning performance in tasks such as disease classification. Domain harmonization is thus a critical research focus. Recent approaches encode brain images $\boldsymbol{x}$ into a low-dimensional latent space $\boldsymbol{z}$, then disentangle it into $\boldsymbol{z_u}$ (domain-invariant) and $\boldsymbol{z_d}$ (domain-specific), achieving strong results. However, these methods often lack interpretability$-$an essential requirement in medical applications$-$leaving practical issues unresolved. We propose Pseudo-Linear-Style Encoder Adversarial Domain Adaptation (PL-SE-ADA), a general framework for domain harmonization and interpretable representation learning that preserves disease-relevant information in brain MR images. PL-SE-ADA includes two encoders $f_E$ and $f_{SE}$ to extract $\boldsymbol{z_u}$ and $\boldsymbol{z_d}$, a decoder to reconstruct the image $f_D$, and a domain predictor $g_D$. Beyond adversarial training between the encoder and domain predictor, the model learns to reconstruct the input image $\boldsymbol{x}$ by summing reconstructions from $\boldsymbol{z_u}$ and $\boldsymbol{z_d}$, ensuring both harmonization and informativeness. Compared to prior methods, PL-SE-ADA achieves equal or better performance in image reconstruction, disease classification, and domain recognition. It also enables visualization of both domain-independent brain features and domain-specific components, offering high interpretability across the entire framework.
SciGA: A Comprehensive Dataset for Designing Graphical Abstracts in Academic Papers
Jul 03, 2025Abstract:Graphical Abstracts (GAs) play a crucial role in visually conveying the key findings of scientific papers. While recent research has increasingly incorporated visual materials such as Figure 1 as de facto GAs, their potential to enhance scientific communication remains largely unexplored. Moreover, designing effective GAs requires advanced visualization skills, creating a barrier to their widespread adoption. To tackle these challenges, we introduce SciGA-145k, a large-scale dataset comprising approximately 145,000 scientific papers and 1.14 million figures, explicitly designed for supporting GA selection and recommendation as well as facilitating research in automated GA generation. As a preliminary step toward GA design support, we define two tasks: 1) Intra-GA recommendation, which identifies figures within a given paper that are well-suited to serve as GAs, and 2) Inter-GA recommendation, which retrieves GAs from other papers to inspire the creation of new GAs. We provide reasonable baseline models for these tasks. Furthermore, we propose Confidence Adjusted top-1 ground truth Ratio (CAR), a novel recommendation metric that offers a fine-grained analysis of model behavior. CAR addresses limitations in traditional ranking-based metrics by considering cases where multiple figures within a paper, beyond the explicitly labeled GA, may also serve as GAs. By unifying these tasks and metrics, our SciGA-145k establishes a foundation for advancing visual scientific communication while contributing to the development of AI for Science.
iCBIR-Sli: Interpretable Content-Based Image Retrieval with 2D Slice Embeddings
Jan 03, 2025


Abstract:Current methods for searching brain MR images rely on text-based approaches, highlighting a significant need for content-based image retrieval (CBIR) systems. Directly applying 3D brain MR images to machine learning models offers the benefit of effectively learning the brain's structure; however, building the generalized model necessitates a large amount of training data. While models that consider depth direction and utilize continuous 2D slices have demonstrated success in segmentation and classification tasks involving 3D data, concerns remain. Specifically, using general 2D slices may lead to the oversight of pathological features and discontinuities in depth direction information. Furthermore, to the best of the authors' knowledge, there have been no attempts to develop a practical CBIR system that preserves the entire brain's structural information. In this study, we propose an interpretable CBIR method for brain MR images, named iCBIR-Sli (Interpretable CBIR with 2D Slice Embedding), which, for the first time globally, utilizes a series of 2D slices. iCBIR-Sli addresses the challenges associated with using 2D slices by effectively aggregating slice information, thereby achieving low-dimensional representations with high completeness, usability, robustness, and interoperability, which are qualities essential for effective CBIR. In retrieval evaluation experiments utilizing five publicly available brain MR datasets (ADNI2/3, OASIS3/4, AIBL) for Alzheimer's disease and cognitively normal, iCBIR-Sli demonstrated top-1 retrieval performance (macro F1 = 0.859), comparable to existing deep learning models explicitly designed for classification, without the need for an external classifier. Additionally, the method provided high interpretability by clearly identifying the brain regions indicative of the searched-for disease.
* 8 pages, 2 figures. Accepted at the SPIE Medical Imaging
Domain-invariant feature learning in brain MR imaging for content-based image retrieval
Jan 02, 2025Abstract:When conducting large-scale studies that collect brain MR images from multiple facilities, the impact of differences in imaging equipment and protocols at each site cannot be ignored, and this domain gap has become a significant issue in recent years. In this study, we propose a new low-dimensional representation (LDR) acquisition method called style encoder adversarial domain adaptation (SE-ADA) to realize content-based image retrieval (CBIR) of brain MR images. SE-ADA reduces domain differences while preserving pathological features by separating domain-specific information from LDR and minimizing domain differences using adversarial learning. In evaluation experiments comparing SE-ADA with recent domain harmonization methods on eight public brain MR datasets (ADNI1/2/3, OASIS1/2/3/4, PPMI), SE-ADA effectively removed domain information while preserving key aspects of the original brain structure and demonstrated the highest disease search accuracy.
* 6 pages, 1 figures. Accepted at the SPIE Medical Imaging 2025
DDD: Discriminative Difficulty Distance for plant disease diagnosis
Jan 01, 2025



Abstract:Recent studies on plant disease diagnosis using machine learning (ML) have highlighted concerns about the overestimated diagnostic performance due to inappropriate data partitioning, where training and test datasets are derived from the same source (domain). Plant disease diagnosis presents a challenging classification task, characterized by its fine-grained nature, vague symptoms, and the extensive variability of image features within each domain. In this study, we propose the concept of Discriminative Difficulty Distance (DDD), a novel metric designed to quantify the domain gap between training and test datasets while assessing the classification difficulty of test data. DDD provides a valuable tool for identifying insufficient diversity in training data, thus supporting the development of more diverse and robust datasets. We investigated multiple image encoders trained on different datasets and examined whether the distances between datasets, measured using low-dimensional representations generated by the encoders, are suitable as a DDD metric. The study utilized 244,063 plant disease images spanning four crops and 34 disease classes collected from 27 domains. As a result, we demonstrated that even if the test images are from different crops or diseases than those used to train the encoder, incorporating them allows the construction of a distance measure for a dataset that strongly correlates with the difficulty of diagnosis indicated by the disease classifier developed independently. Compared to the base encoder, pre-trained only on ImageNet21K, the correlation higher by 0.106 to 0.485, reaching a maximum of 0.909.
Few-shot Metric Domain Adaptation: Practical Learning Strategies for an Automated Plant Disease Diagnosis
Dec 25, 2024



Abstract:Numerous studies have explored image-based automated systems for plant disease diagnosis, demonstrating impressive diagnostic capabilities. However, recent large-scale analyses have revealed a critical limitation: that the diagnostic capability suffers significantly when validated on images captured in environments (domains) differing from those used during training. This shortfall stems from the inherently limited dataset size and the diverse manifestation of disease symptoms, combined with substantial variations in cultivation environments and imaging conditions, such as equipment and composition. These factors lead to insufficient variety in training data, ultimately constraining the system's robustness and generalization. To address these challenges, we propose Few-shot Metric Domain Adaptation (FMDA), a flexible and effective approach for enhancing diagnostic accuracy in practical systems, even when only limited target data is available. FMDA reduces domain discrepancies by introducing a constraint to the diagnostic model that minimizes the "distance" between feature spaces of source (training) data and target data with limited samples. FMDA is computationally efficient, requiring only basic feature distance calculations and backpropagation, and can be seamlessly integrated into any machine learning (ML) pipeline. In large-scale experiments, involving 223,015 leaf images across 20 fields and 3 crop species, FMDA achieved F1 score improvements of 11.1 to 29.3 points compared to cases without target data, using only 10 images per disease from the target domain. Moreover, FMDA consistently outperformed fine-tuning methods utilizing the same data, with an average improvement of 8.5 points.
Extended Japanese Commonsense Morality Dataset with Masked Token and Label Enhancement
Oct 12, 2024Abstract:Rapid advancements in artificial intelligence (AI) have made it crucial to integrate moral reasoning into AI systems. However, existing models and datasets often overlook regional and cultural differences. To address this shortcoming, we have expanded the JCommonsenseMorality (JCM) dataset, the only publicly available dataset focused on Japanese morality. The Extended JCM (eJCM) has grown from the original 13,975 sentences to 31,184 sentences using our proposed sentence expansion method called Masked Token and Label Enhancement (MTLE). MTLE selectively masks important parts of sentences related to moral judgment and replaces them with alternative expressions generated by a large language model (LLM), while re-assigning appropriate labels. The model trained using our eJCM achieved an F1 score of 0.857, higher than the scores for the original JCM (0.837), ChatGPT one-shot classification (0.841), and data augmented using AugGPT, a state-of-the-art augmentation method (0.850). Specifically, in complex moral reasoning tasks unique to Japanese culture, the model trained with eJCM showed a significant improvement in performance (increasing from 0.681 to 0.756) and achieved a performance close to that of GPT-4 Turbo (0.787). These results demonstrate the validity of the eJCM dataset and the importance of developing models and datasets that consider the cultural context.
Hierarchical Object Detection and Recognition Framework for Practical Plant Disease Diagnosis
Jul 25, 2024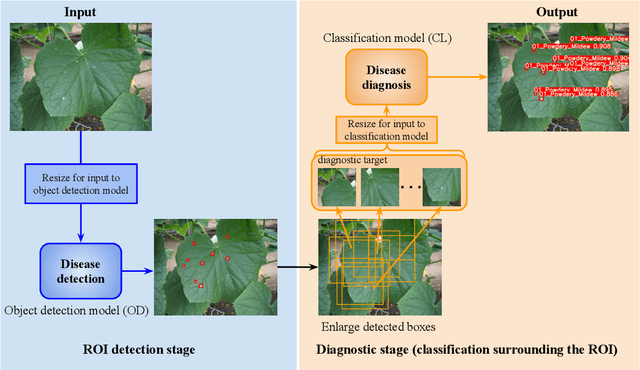
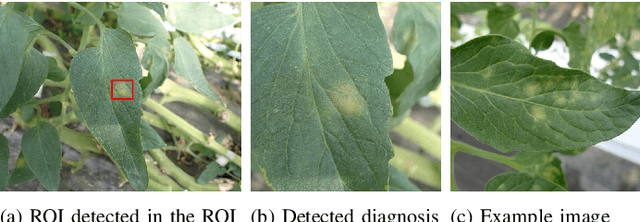

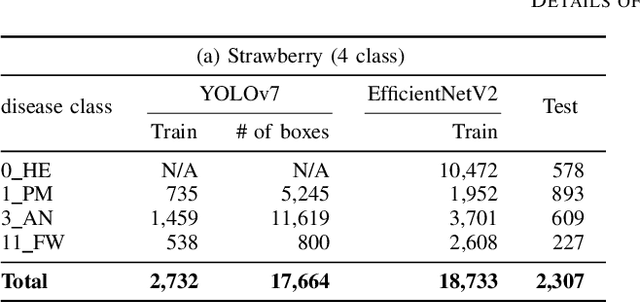
Abstract:Recently, object detection methods (OD; e.g., YOLO-based models) have been widely utilized in plant disease diagnosis. These methods demonstrate robustness to distance variations and excel at detecting small lesions compared to classification methods (CL; e.g., CNN models). However, there are issues such as low diagnostic performance for hard-to-detect diseases and high labeling costs. Additionally, since healthy cases cannot be explicitly trained, there is a risk of false positives. We propose the Hierarchical object detection and recognition framework (HODRF), a sophisticated and highly integrated two-stage system that combines the strengths of both OD and CL for plant disease diagnosis. In the first stage, HODRF uses OD to identify regions of interest (ROIs) without specifying the disease. In the second stage, CL diagnoses diseases surrounding the ROIs. HODRF offers several advantages: (1) Since OD detects only one type of ROI, HODRF can detect diseases with limited training images by leveraging its ability to identify other lesions. (2) While OD over-detects healthy cases, HODRF significantly reduces these errors by using CL in the second stage. (3) CL's accuracy improves in HODRF as it identifies diagnostic targets given as ROIs, making it less vulnerable to size changes. (4) HODRF benefits from CL's lower annotation costs, allowing it to learn from a larger number of images. We implemented HODRF using YOLOv7 for OD and EfficientNetV2 for CL and evaluated its performance on a large-scale dataset (4 crops, 20 diseased and healthy classes, 281K images). HODRF outperformed YOLOv7 alone by 5.8 to 21.5 points on healthy data and 0.6 to 7.5 points on macro F1 scores, and it improved macro F1 by 1.1 to 7.2 points over EfficientNetV2.
Investigation to answer three key questions concerning plant pest identification and development of a practical identification framework
Jul 25, 2024Abstract:The development of practical and robust automated diagnostic systems for identifying plant pests is crucial for efficient agricultural production. In this paper, we first investigate three key research questions (RQs) that have not been addressed thus far in the field of image-based plant pest identification. Based on the knowledge gained, we then develop an accurate, robust, and fast plant pest identification framework using 334K images comprising 78 combinations of four plant portions (the leaf front, leaf back, fruit, and flower of cucumber, tomato, strawberry, and eggplant) and 20 pest species captured at 27 farms. The results reveal the following. (1) For an appropriate evaluation of the model, the test data should not include images of the field from which the training images were collected, or other considerations to increase the diversity of the test set should be taken into account. (2) Pre-extraction of ROIs, such as leaves and fruits, helps to improve identification accuracy. (3) Integration of closely related species using the same control methods and cross-crop training methods for the same pests, are effective. Our two-stage plant pest identification framework, enabling ROI detection and convolutional neural network (CNN)-based identification, achieved a highly practical performance of 91.0% and 88.5% in mean accuracy and macro F1 score, respectively, for 12,223 instances of test data of 21 classes collected from unseen fields, where 25 classes of images from 318,971 samples were used for training; the average identification time was 476 ms/image.
* 40 pages, 10 figures
Majority or Minority: Data Imbalance Learning Method for Named Entity Recognition
Jan 21, 2024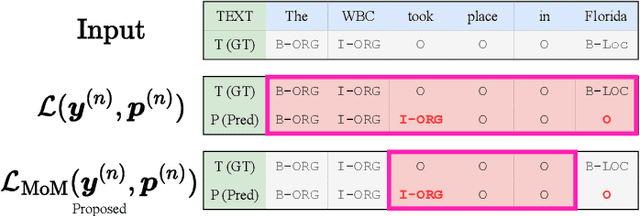
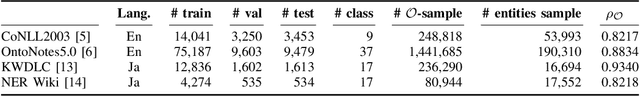
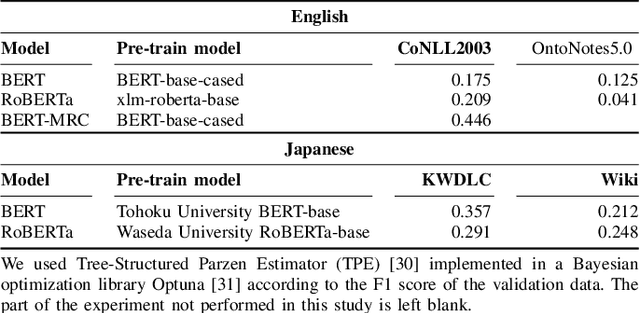
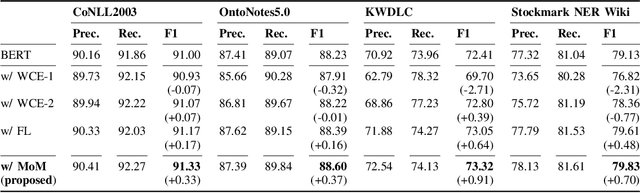
Abstract:Data imbalance presents a significant challenge in various machine learning (ML) tasks, particularly named entity recognition (NER) within natural language processing (NLP). NER exhibits a data imbalance with a long-tail distribution, featuring numerous minority classes (i.e., entity classes) and a single majority class (i.e., O-class). The imbalance leads to the misclassifications of the entity classes as the O-class. To tackle the imbalance, we propose a simple and effective learning method, named majority or minority (MoM) learning. MoM learning incorporates the loss computed only for samples whose ground truth is the majority class (i.e., the O-class) into the loss of the conventional ML model. Evaluation experiments on four NER datasets (Japanese and English) showed that MoM learning improves prediction performance of the minority classes, without sacrificing the performance of the majority class and is more effective than widely known and state-of-the-art methods. We also evaluated MoM learning using frameworks as sequential labeling and machine reading comprehension, which are commonly used in NER. Furthermore, MoM learning has achieved consistent performance improvements regardless of language, model, or framework.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge