Kenichi Oishi
Acquisition of interpretable domain information during brain MR image harmonization for content-based image retrieval
Oct 16, 2025Abstract:Medical images like MR scans often show domain shifts across imaging sites due to scanner and protocol differences, which degrade machine learning performance in tasks such as disease classification. Domain harmonization is thus a critical research focus. Recent approaches encode brain images $\boldsymbol{x}$ into a low-dimensional latent space $\boldsymbol{z}$, then disentangle it into $\boldsymbol{z_u}$ (domain-invariant) and $\boldsymbol{z_d}$ (domain-specific), achieving strong results. However, these methods often lack interpretability$-$an essential requirement in medical applications$-$leaving practical issues unresolved. We propose Pseudo-Linear-Style Encoder Adversarial Domain Adaptation (PL-SE-ADA), a general framework for domain harmonization and interpretable representation learning that preserves disease-relevant information in brain MR images. PL-SE-ADA includes two encoders $f_E$ and $f_{SE}$ to extract $\boldsymbol{z_u}$ and $\boldsymbol{z_d}$, a decoder to reconstruct the image $f_D$, and a domain predictor $g_D$. Beyond adversarial training between the encoder and domain predictor, the model learns to reconstruct the input image $\boldsymbol{x}$ by summing reconstructions from $\boldsymbol{z_u}$ and $\boldsymbol{z_d}$, ensuring both harmonization and informativeness. Compared to prior methods, PL-SE-ADA achieves equal or better performance in image reconstruction, disease classification, and domain recognition. It also enables visualization of both domain-independent brain features and domain-specific components, offering high interpretability across the entire framework.
iCBIR-Sli: Interpretable Content-Based Image Retrieval with 2D Slice Embeddings
Jan 03, 2025


Abstract:Current methods for searching brain MR images rely on text-based approaches, highlighting a significant need for content-based image retrieval (CBIR) systems. Directly applying 3D brain MR images to machine learning models offers the benefit of effectively learning the brain's structure; however, building the generalized model necessitates a large amount of training data. While models that consider depth direction and utilize continuous 2D slices have demonstrated success in segmentation and classification tasks involving 3D data, concerns remain. Specifically, using general 2D slices may lead to the oversight of pathological features and discontinuities in depth direction information. Furthermore, to the best of the authors' knowledge, there have been no attempts to develop a practical CBIR system that preserves the entire brain's structural information. In this study, we propose an interpretable CBIR method for brain MR images, named iCBIR-Sli (Interpretable CBIR with 2D Slice Embedding), which, for the first time globally, utilizes a series of 2D slices. iCBIR-Sli addresses the challenges associated with using 2D slices by effectively aggregating slice information, thereby achieving low-dimensional representations with high completeness, usability, robustness, and interoperability, which are qualities essential for effective CBIR. In retrieval evaluation experiments utilizing five publicly available brain MR datasets (ADNI2/3, OASIS3/4, AIBL) for Alzheimer's disease and cognitively normal, iCBIR-Sli demonstrated top-1 retrieval performance (macro F1 = 0.859), comparable to existing deep learning models explicitly designed for classification, without the need for an external classifier. Additionally, the method provided high interpretability by clearly identifying the brain regions indicative of the searched-for disease.
* 8 pages, 2 figures. Accepted at the SPIE Medical Imaging
Domain-invariant feature learning in brain MR imaging for content-based image retrieval
Jan 02, 2025Abstract:When conducting large-scale studies that collect brain MR images from multiple facilities, the impact of differences in imaging equipment and protocols at each site cannot be ignored, and this domain gap has become a significant issue in recent years. In this study, we propose a new low-dimensional representation (LDR) acquisition method called style encoder adversarial domain adaptation (SE-ADA) to realize content-based image retrieval (CBIR) of brain MR images. SE-ADA reduces domain differences while preserving pathological features by separating domain-specific information from LDR and minimizing domain differences using adversarial learning. In evaluation experiments comparing SE-ADA with recent domain harmonization methods on eight public brain MR datasets (ADNI1/2/3, OASIS1/2/3/4, PPMI), SE-ADA effectively removed domain information while preserving key aspects of the original brain structure and demonstrated the highest disease search accuracy.
* 6 pages, 1 figures. Accepted at the SPIE Medical Imaging 2025
Loc-VAE: Learning Structurally Localized Representation from 3D Brain MR Images for Content-Based Image Retrieval
Oct 02, 2022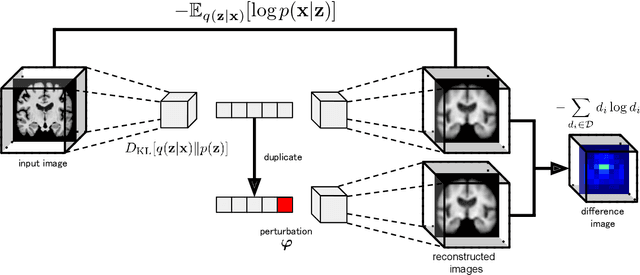
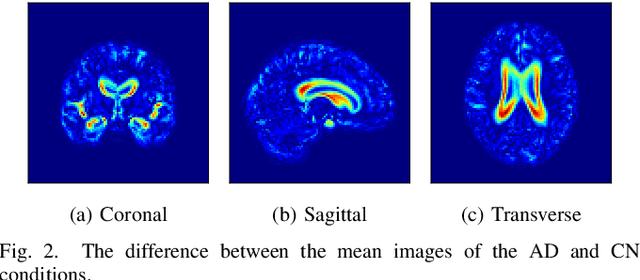
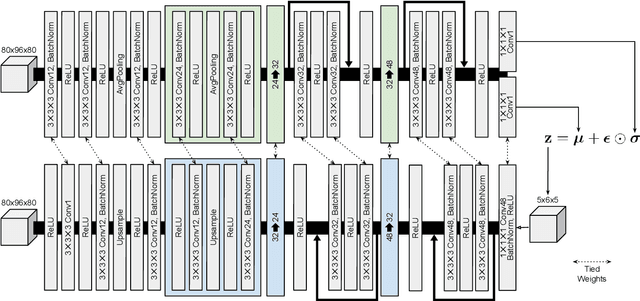
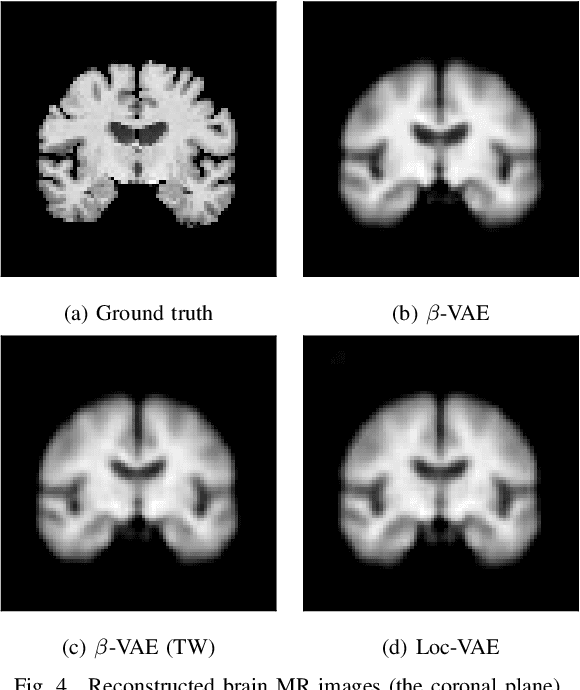
Abstract:Content-based image retrieval (CBIR) systems are an emerging technology that supports reading and interpreting medical images. Since 3D brain MR images are high dimensional, dimensionality reduction is necessary for CBIR using machine learning techniques. In addition, for a reliable CBIR system, each dimension in the resulting low-dimensional representation must be associated with a neurologically interpretable region. We propose a localized variational autoencoder (Loc-VAE) that provides neuroanatomically interpretable low-dimensional representation from 3D brain MR images for clinical CBIR. Loc-VAE is based on $\beta$-VAE with the additional constraint that each dimension of the low-dimensional representation corresponds to a local region of the brain. The proposed Loc-VAE is capable of acquiring representation that preserves disease features and is highly localized, even under high-dimensional compression ratios (4096:1). The low-dimensional representation obtained by Loc-VAE improved the locality measure of each dimension by 4.61 points compared to naive $\beta$-VAE, while maintaining comparable brain reconstruction capability and information about the diagnosis of Alzheimer's disease.
Disease-oriented image embedding with pseudo-scanner standardization for content-based image retrieval on 3D brain MRI
Aug 14, 2021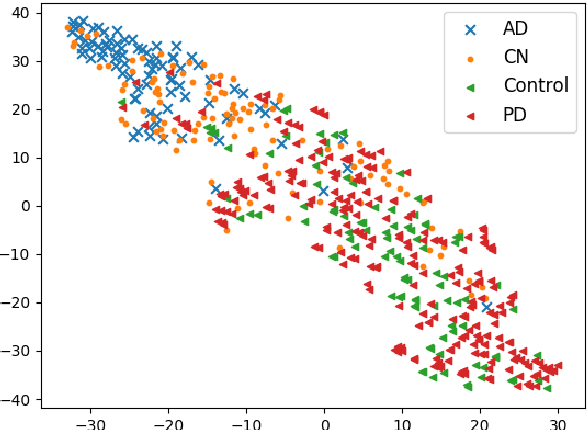
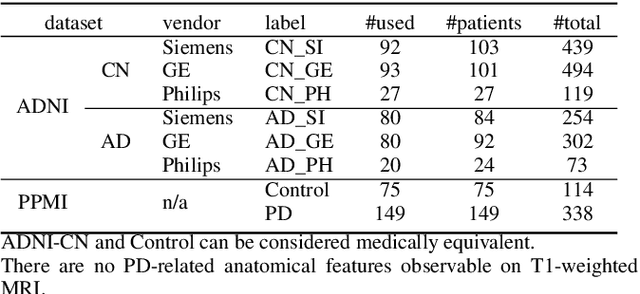
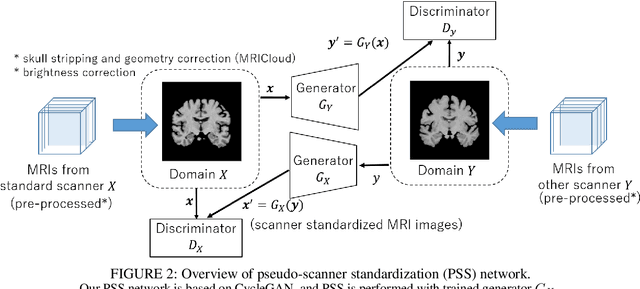
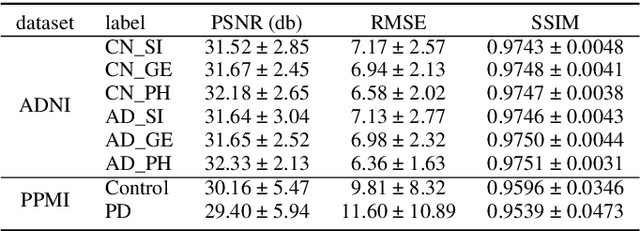
Abstract:To build a robust and practical content-based image retrieval (CBIR) system that is applicable to a clinical brain MRI database, we propose a new framework -- Disease-oriented image embedding with pseudo-scanner standardization (DI-PSS) -- that consists of two core techniques, data harmonization and a dimension reduction algorithm. Our DI-PSS uses skull stripping and CycleGAN-based image transformations that map to a standard brain followed by transformation into a brain image taken with a given reference scanner. Then, our 3D convolutioinal autoencoders (3D-CAE) with deep metric learning acquires a low-dimensional embedding that better reflects the characteristics of the disease. The effectiveness of our proposed framework was tested on the T1-weighted MRIs selected from the Alzheimer's Disease Neuroimaging Initiative and the Parkinson's Progression Markers Initiative. We confirmed that our PSS greatly reduced the variability of low-dimensional embeddings caused by different scanner and datasets. Compared with the baseline condition, our PSS reduced the variability in the distance from Alzheimer's disease (AD) to clinically normal (CN) and Parkinson disease (PD) cases by 15.8-22.6% and 18.0-29.9%, respectively. These properties allow DI-PSS to generate lower dimensional representations that are more amenable to disease classification. In AD and CN classification experiments based on spectral clustering, PSS improved the average accuracy and macro-F1 by 6.2% and 10.7%, respectively. Given the potential of the DI-PSS for harmonizing images scanned by MRI scanners that were not used to scan the training data, we expect that the DI-PSS is suitable for application to a large number of legacy MRIs scanned in heterogeneous environments.
Efficient feature embedding of 3D brain MRI images for content-based image retrieval with deep metric learning
Dec 04, 2019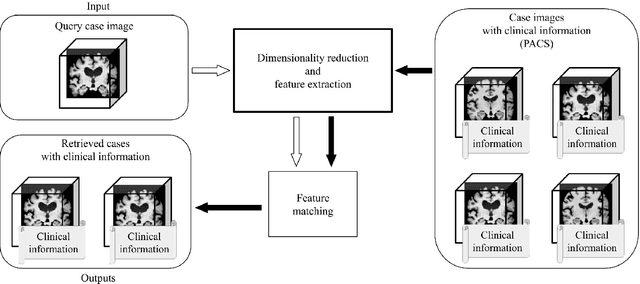
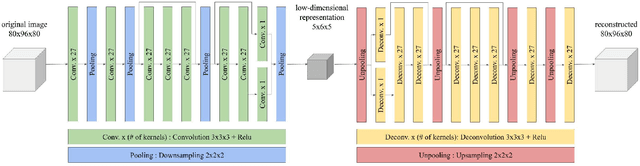


Abstract:Increasing numbers of MRI brain scans, improvements in image resolution, and advancements in MRI acquisition technology are causing significant increases in the demand for and burden on radiologists' efforts in terms of reading and interpreting brain MRIs. Content-based image retrieval (CBIR) is an emerging technology for reducing this burden by supporting the reading of medical images. High dimensionality is a major challenge in developing a CBIR system that is applicable for 3D brain MRIs. In this study, we propose a system called disease-oriented data concentration with metric learning (DDCML). In DDCML, we introduce deep metric learning to a 3D convolutional autoencoder (CAE). Our proposed DDCML scheme achieves a high dimensional compression rate (4096:1) while preserving the disease-related anatomical features that are important for medical image classification. The low-dimensional representation obtained by DDCML improved the clustering performance by 29.1\% compared to plain 3D-CAE in terms of discriminating Alzheimer's disease patients from healthy subjects, and successfully reproduced the relationships of the severity of disease categories that were not included in the training.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge