Kumpei Ikuta
Loc-VAE: Learning Structurally Localized Representation from 3D Brain MR Images for Content-Based Image Retrieval
Oct 02, 2022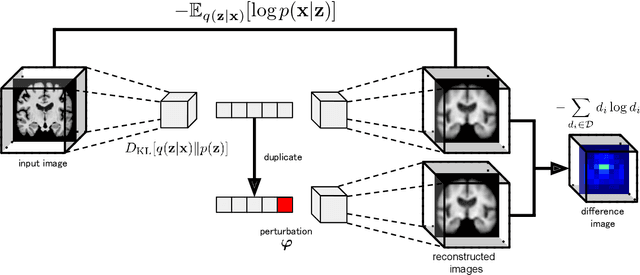
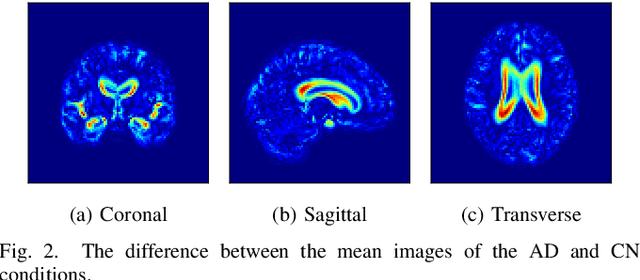
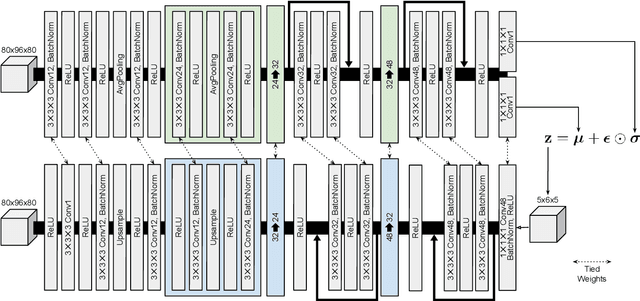
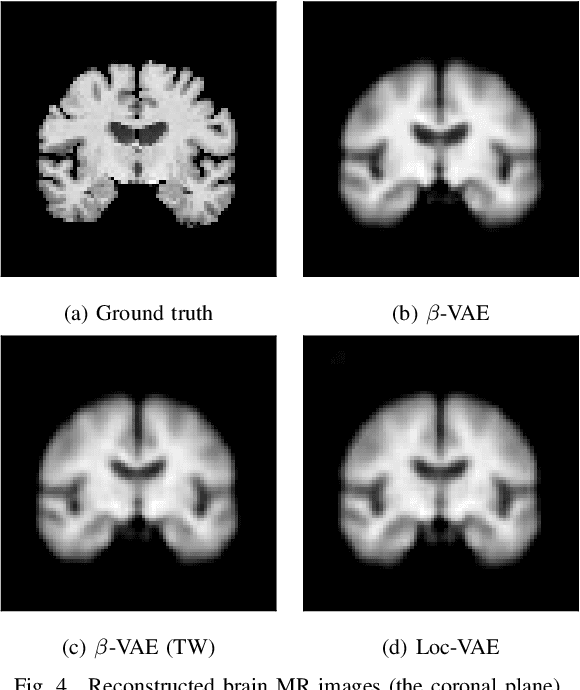
Abstract:Content-based image retrieval (CBIR) systems are an emerging technology that supports reading and interpreting medical images. Since 3D brain MR images are high dimensional, dimensionality reduction is necessary for CBIR using machine learning techniques. In addition, for a reliable CBIR system, each dimension in the resulting low-dimensional representation must be associated with a neurologically interpretable region. We propose a localized variational autoencoder (Loc-VAE) that provides neuroanatomically interpretable low-dimensional representation from 3D brain MR images for clinical CBIR. Loc-VAE is based on $\beta$-VAE with the additional constraint that each dimension of the low-dimensional representation corresponds to a local region of the brain. The proposed Loc-VAE is capable of acquiring representation that preserves disease features and is highly localized, even under high-dimensional compression ratios (4096:1). The low-dimensional representation obtained by Loc-VAE improved the locality measure of each dimension by 4.61 points compared to naive $\beta$-VAE, while maintaining comparable brain reconstruction capability and information about the diagnosis of Alzheimer's disease.
Disease-oriented image embedding with pseudo-scanner standardization for content-based image retrieval on 3D brain MRI
Aug 14, 2021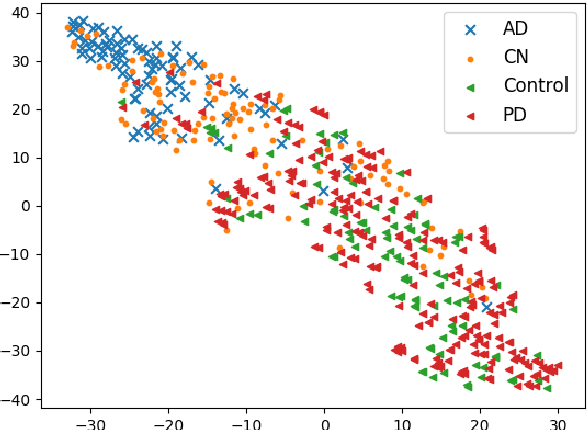
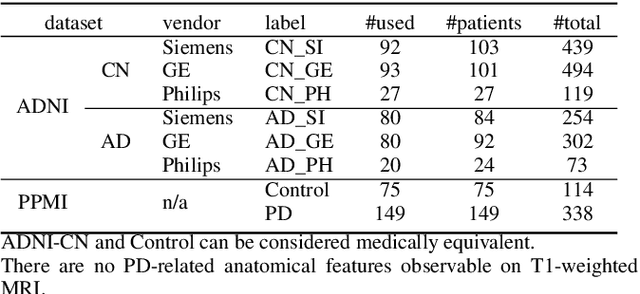
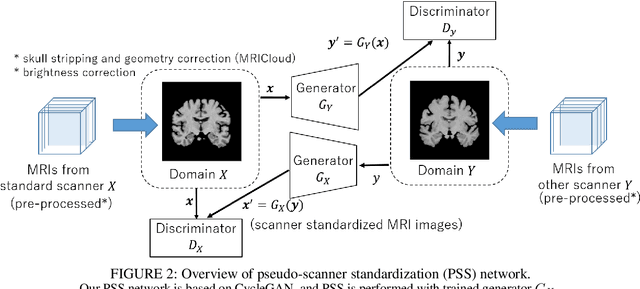
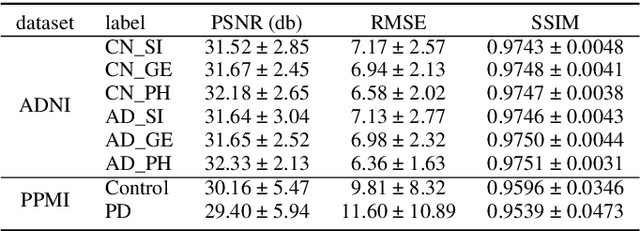
Abstract:To build a robust and practical content-based image retrieval (CBIR) system that is applicable to a clinical brain MRI database, we propose a new framework -- Disease-oriented image embedding with pseudo-scanner standardization (DI-PSS) -- that consists of two core techniques, data harmonization and a dimension reduction algorithm. Our DI-PSS uses skull stripping and CycleGAN-based image transformations that map to a standard brain followed by transformation into a brain image taken with a given reference scanner. Then, our 3D convolutioinal autoencoders (3D-CAE) with deep metric learning acquires a low-dimensional embedding that better reflects the characteristics of the disease. The effectiveness of our proposed framework was tested on the T1-weighted MRIs selected from the Alzheimer's Disease Neuroimaging Initiative and the Parkinson's Progression Markers Initiative. We confirmed that our PSS greatly reduced the variability of low-dimensional embeddings caused by different scanner and datasets. Compared with the baseline condition, our PSS reduced the variability in the distance from Alzheimer's disease (AD) to clinically normal (CN) and Parkinson disease (PD) cases by 15.8-22.6% and 18.0-29.9%, respectively. These properties allow DI-PSS to generate lower dimensional representations that are more amenable to disease classification. In AD and CN classification experiments based on spectral clustering, PSS improved the average accuracy and macro-F1 by 6.2% and 10.7%, respectively. Given the potential of the DI-PSS for harmonizing images scanned by MRI scanners that were not used to scan the training data, we expect that the DI-PSS is suitable for application to a large number of legacy MRIs scanned in heterogeneous environments.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge