Loc-VAE: Learning Structurally Localized Representation from 3D Brain MR Images for Content-Based Image Retrieval
Paper and Code
Oct 02, 2022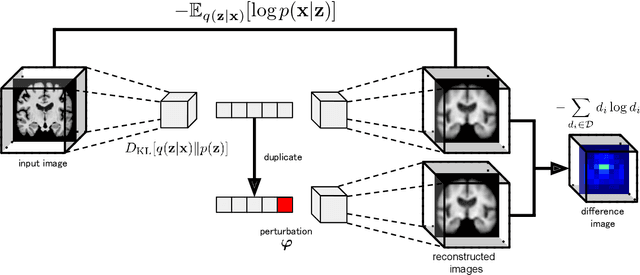
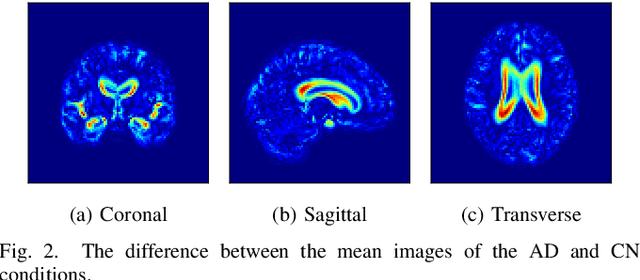
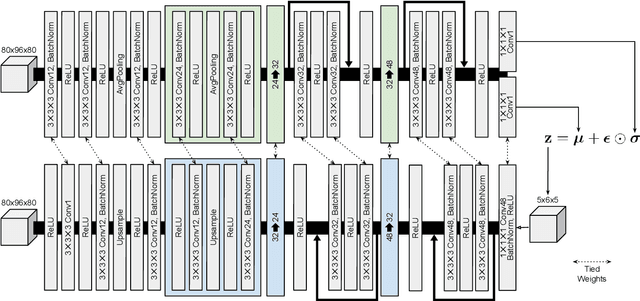
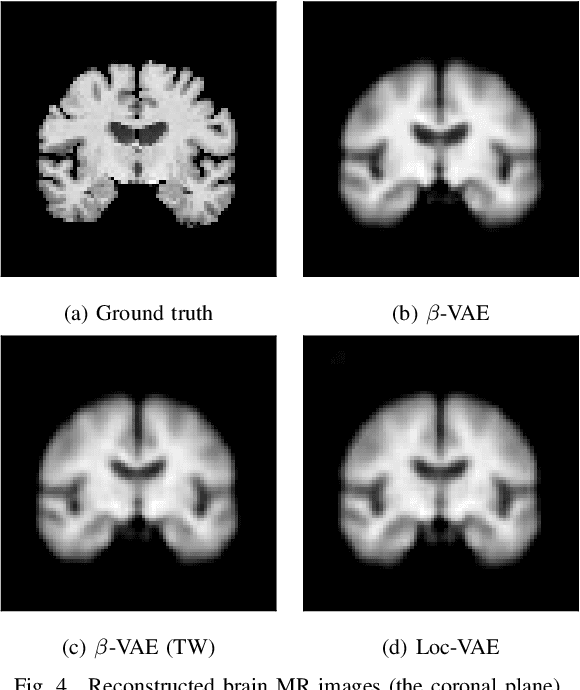
Content-based image retrieval (CBIR) systems are an emerging technology that supports reading and interpreting medical images. Since 3D brain MR images are high dimensional, dimensionality reduction is necessary for CBIR using machine learning techniques. In addition, for a reliable CBIR system, each dimension in the resulting low-dimensional representation must be associated with a neurologically interpretable region. We propose a localized variational autoencoder (Loc-VAE) that provides neuroanatomically interpretable low-dimensional representation from 3D brain MR images for clinical CBIR. Loc-VAE is based on $\beta$-VAE with the additional constraint that each dimension of the low-dimensional representation corresponds to a local region of the brain. The proposed Loc-VAE is capable of acquiring representation that preserves disease features and is highly localized, even under high-dimensional compression ratios (4096:1). The low-dimensional representation obtained by Loc-VAE improved the locality measure of each dimension by 4.61 points compared to naive $\beta$-VAE, while maintaining comparable brain reconstruction capability and information about the diagnosis of Alzheimer's disease.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge