Hierarchical Object Detection and Recognition Framework for Practical Plant Disease Diagnosis
Paper and Code
Jul 25, 2024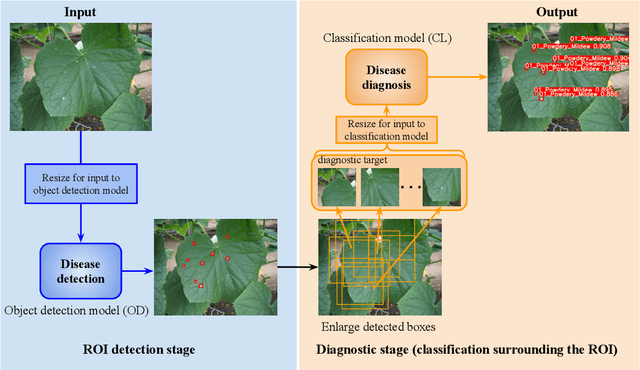
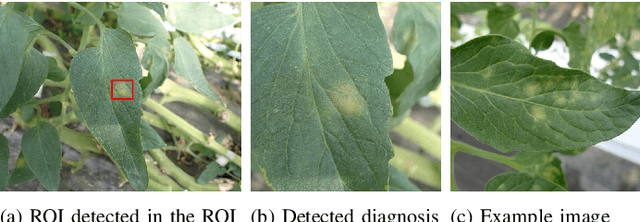

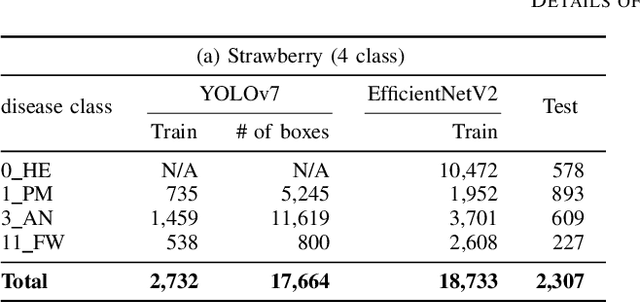
Recently, object detection methods (OD; e.g., YOLO-based models) have been widely utilized in plant disease diagnosis. These methods demonstrate robustness to distance variations and excel at detecting small lesions compared to classification methods (CL; e.g., CNN models). However, there are issues such as low diagnostic performance for hard-to-detect diseases and high labeling costs. Additionally, since healthy cases cannot be explicitly trained, there is a risk of false positives. We propose the Hierarchical object detection and recognition framework (HODRF), a sophisticated and highly integrated two-stage system that combines the strengths of both OD and CL for plant disease diagnosis. In the first stage, HODRF uses OD to identify regions of interest (ROIs) without specifying the disease. In the second stage, CL diagnoses diseases surrounding the ROIs. HODRF offers several advantages: (1) Since OD detects only one type of ROI, HODRF can detect diseases with limited training images by leveraging its ability to identify other lesions. (2) While OD over-detects healthy cases, HODRF significantly reduces these errors by using CL in the second stage. (3) CL's accuracy improves in HODRF as it identifies diagnostic targets given as ROIs, making it less vulnerable to size changes. (4) HODRF benefits from CL's lower annotation costs, allowing it to learn from a larger number of images. We implemented HODRF using YOLOv7 for OD and EfficientNetV2 for CL and evaluated its performance on a large-scale dataset (4 crops, 20 diseased and healthy classes, 281K images). HODRF outperformed YOLOv7 alone by 5.8 to 21.5 points on healthy data and 0.6 to 7.5 points on macro F1 scores, and it improved macro F1 by 1.1 to 7.2 points over EfficientNetV2.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge