Kaiyuan Yang
PowerGenie: Analytically-Guided Evolutionary Discovery of Superior Reconfigurable Power Converters
Jan 29, 2026Abstract:Discovering superior circuit topologies requires navigating an exponentially large design space-a challenge traditionally reserved for human experts. Existing AI methods either select from predefined templates or generate novel topologies at a limited scale without rigorous verification, leaving large-scale performance-driven discovery underexplored. We present PowerGenie, a framework for automated discovery of higher-performance reconfigurable power converters at scale. PowerGenie introduces: (1) an automated analytical framework that determines converter functionality and theoretical performance limits without component sizing or SPICE simulation, and (2) an evolutionary finetuning method that co-evolves a generative model with its training distribution through fitness selection and uniqueness verification. Unlike existing methods that suffer from mode collapse and overfitting, our approach achieves higher syntax validity, function validity, novelty rate, and figure-of-merit (FoM). PowerGenie discovers a novel 8-mode reconfigurable converter with 23% higher FoM than the best training topology. SPICE simulations confirm average absolute efficiency gains of 10% across 8 modes and up to 17% at a single mode. Code is available at https://github.com/xz-group/PowerGenie.
Compressive Sensing Photoacoustic Imaging Receiver with Matrix-Vector-Multiplication SAR ADC
Nov 09, 2025Abstract:Wearable photoacoustic imaging devices hold great promise for continuous health monitoring and point-of-care diagnostics. However, the large data volume generated by high-density transducer arrays presents a major challenge for realizing compact and power-efficient wearable systems. This paper presents a photoacoustic imaging receiver (RX) that embeds compressive sensing directly into the hardware to address this bottleneck. The RX integrates 16 AFEs and four matrix-vector-multiplication (MVM) SAR ADCs that perform energy- and area-efficient analog-domain compression. The architecture achieves a 4-8x reduction in output data rate while preserving low-loss full-array information. The MVM SAR ADC executes passive and accurate MVM using user-defined programmable ternary weights. Two signal reconstruction methods are implemented: (1) an optimization approach using the fast iterative shrinkage-thresholding algorithm, and (2) a learning-based approach employing implicit neural representation. Fabricated in 65 nm CMOS, the chip achieves an ADC's SNDR of 57.5 dB at 20.41 MS/s, with an AFE input-referred noise of 3.5 nV/sqrt(Hz). MVM linearity measurements show R^2 > 0.999 across a wide range of weights and input amplitudes. The system is validated through phantom imaging experiments, demonstrating high-fidelity image reconstruction under up to 8x compression. The RX consumes 5.83 mW/channel and supports a general ternary-weighted measurement matrix, offering a compelling solution for next-generation miniaturized, wearable PA imaging systems.
Parametric shape models for vessels learned from segmentations via differentiable voxelization
Jul 03, 2025Abstract:Vessels are complex structures in the body that have been studied extensively in multiple representations. While voxelization is the most common of them, meshes and parametric models are critical in various applications due to their desirable properties. However, these representations are typically extracted through segmentations and used disjointly from each other. We propose a framework that joins the three representations under differentiable transformations. By leveraging differentiable voxelization, we automatically extract a parametric shape model of the vessels through shape-to-segmentation fitting, where we learn shape parameters from segmentations without the explicit need for ground-truth shape parameters. The vessel is parametrized as centerlines and radii using cubic B-splines, ensuring smoothness and continuity by construction. Meshes are differentiably extracted from the learned shape parameters, resulting in high-fidelity meshes that can be manipulated post-fit. Our method can accurately capture the geometry of complex vessels, as demonstrated by the volumetric fits in experiments on aortas, aneurysms, and brain vessels.
FlexDuo: A Pluggable System for Enabling Full-Duplex Capabilities in Speech Dialogue Systems
Feb 19, 2025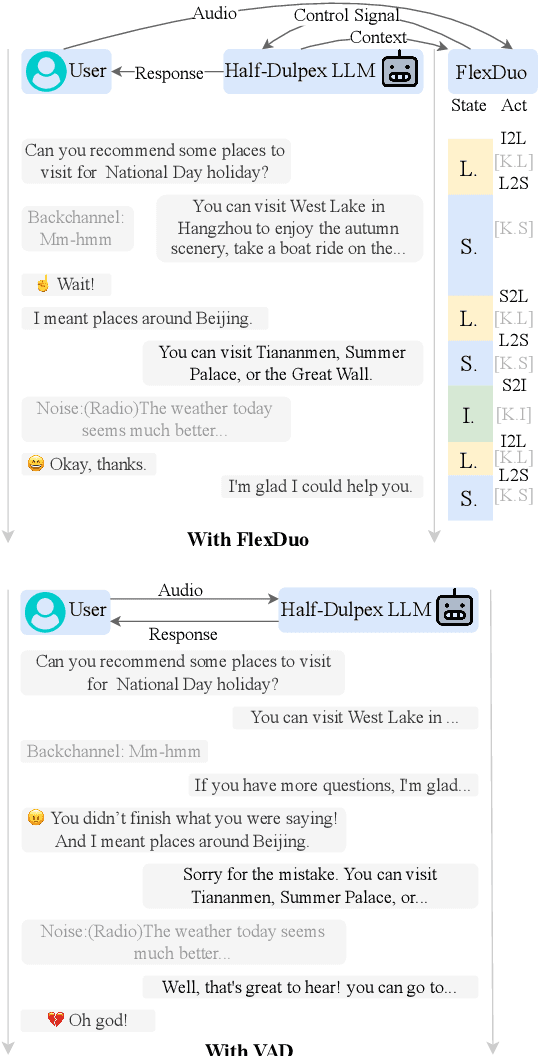

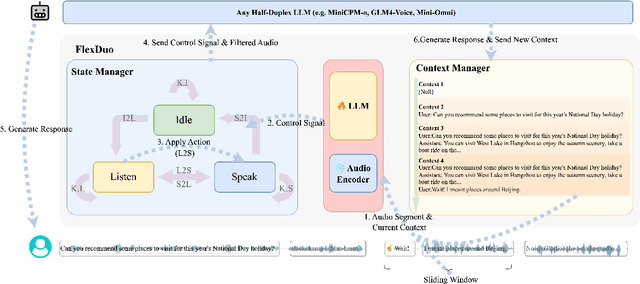

Abstract:Full-Duplex Speech Dialogue Systems (Full-Duplex SDS) have significantly enhanced the naturalness of human-machine interaction by enabling real-time bidirectional communication. However, existing approaches face challenges such as difficulties in independent module optimization and contextual noise interference due to highly coupled architectural designs and oversimplified binary state modeling. This paper proposes FlexDuo, a flexible full-duplex control module that decouples duplex control from spoken dialogue systems through a plug-and-play architectural design. Furthermore, inspired by human information-filtering mechanisms in conversations, we introduce an explicit Idle state. On one hand, the Idle state filters redundant noise and irrelevant audio to enhance dialogue quality. On the other hand, it establishes a semantic integrity-based buffering mechanism, reducing the risk of mutual interruptions while ensuring accurate response transitions. Experimental results on the Fisher corpus demonstrate that FlexDuo reduces the false interruption rate by 24.9% and improves response accuracy by 7.6% compared to integrated full-duplex dialogue system baselines. It also outperforms voice activity detection (VAD) controlled baseline systems in both Chinese and English dialogue quality. The proposed modular architecture and state-based dialogue model provide a novel technical pathway for building flexible and efficient duplex dialogue systems.
SELMA3D challenge: Self-supervised learning for 3D light-sheet microscopy image segmentation
Jan 07, 2025Abstract:Recent innovations in light sheet microscopy, paired with developments in tissue clearing techniques, enable the 3D imaging of large mammalian tissues with cellular resolution. Combined with the progress in large-scale data analysis, driven by deep learning, these innovations empower researchers to rapidly investigate the morphological and functional properties of diverse biological samples. Segmentation, a crucial preliminary step in the analysis process, can be automated using domain-specific deep learning models with expert-level performance. However, these models exhibit high sensitivity to domain shifts, leading to a significant drop in accuracy when applied to data outside their training distribution. To address this limitation, and inspired by the recent success of self-supervised learning in training generalizable models, we organized the SELMA3D Challenge during the MICCAI 2024 conference. SELMA3D provides a vast collection of light-sheet images from cleared mice and human brains, comprising 35 large 3D images-each with over 1000^3 voxels-and 315 annotated small patches for finetuning, preliminary testing and final testing. The dataset encompasses diverse biological structures, including vessel-like and spot-like structures. Five teams participated in all phases of the challenge, and their proposed methods are reviewed in this paper. Quantitative and qualitative results from most participating teams demonstrate that self-supervised learning on large datasets improves segmentation model performance and generalization. We will continue to support and extend SELMA3D as an inaugural MICCAI challenge focused on self-supervised learning for 3D microscopy image segmentation.
ISLES 2024: The first longitudinal multimodal multi-center real-world dataset in (sub-)acute stroke
Aug 20, 2024
Abstract:Stroke remains a leading cause of global morbidity and mortality, placing a heavy socioeconomic burden. Over the past decade, advances in endovascular reperfusion therapy and the use of CT and MRI imaging for treatment guidance have significantly improved patient outcomes and are now standard in clinical practice. To develop machine learning algorithms that can extract meaningful and reproducible models of brain function for both clinical and research purposes from stroke images - particularly for lesion identification, brain health quantification, and prognosis - large, diverse, and well-annotated public datasets are essential. While only a few datasets with (sub-)acute stroke data were previously available, several large, high-quality datasets have recently been made publicly accessible. However, these existing datasets include only MRI data. In contrast, our dataset is the first to offer comprehensive longitudinal stroke data, including acute CT imaging with angiography and perfusion, follow-up MRI at 2-9 days, as well as acute and longitudinal clinical data up to a three-month outcome. The dataset includes a training dataset of n = 150 and a test dataset of n = 100 scans. Training data is publicly available, while test data will be used exclusively for model validation. We are making this dataset available as part of the 2024 edition of the Ischemic Stroke Lesion Segmentation (ISLES) challenge (https://www.isles-challenge.org/), which continuously aims to establish benchmark methods for acute and sub-acute ischemic stroke lesion segmentation, aiding in creating open stroke imaging datasets and evaluating cutting-edge image processing algorithms.
ISLES'24: Improving final infarct prediction in ischemic stroke using multimodal imaging and clinical data
Aug 20, 2024
Abstract:Accurate estimation of core (irreversibly damaged tissue) and penumbra (salvageable tissue) volumes is essential for ischemic stroke treatment decisions. Perfusion CT, the clinical standard, estimates these volumes but is affected by variations in deconvolution algorithms, implementations, and thresholds. Core tissue expands over time, with growth rates influenced by thrombus location, collateral circulation, and inherent patient-specific factors. Understanding this tissue growth is crucial for determining the need to transfer patients to comprehensive stroke centers, predicting the benefits of additional reperfusion attempts during mechanical thrombectomy, and forecasting final clinical outcomes. This work presents the ISLES'24 challenge, which addresses final post-treatment stroke infarct prediction from pre-interventional acute stroke imaging and clinical data. ISLES'24 establishes a unique 360-degree setting where all feasibly accessible clinical data are available for participants, including full CT acute stroke imaging, sub-acute follow-up MRI, and clinical tabular data. The contributions of this work are two-fold: first, we introduce a standardized benchmarking of final stroke infarct segmentation algorithms through the ISLES'24 challenge; second, we provide insights into infarct segmentation using multimodal imaging and clinical data strategies by identifying outperforming methods on a finely curated dataset. The outputs of this challenge are anticipated to enhance clinical decision-making and improve patient outcome predictions. All ISLES'24 materials, including data, performance evaluation scripts, and leading algorithmic strategies, are available to the research community following \url{https://isles-24.grand-challenge.org/}.
3D Vessel Graph Generation Using Denoising Diffusion
Jul 08, 2024



Abstract:Blood vessel networks, represented as 3D graphs, help predict disease biomarkers, simulate blood flow, and aid in synthetic image generation, relevant in both clinical and pre-clinical settings. However, generating realistic vessel graphs that correspond to an anatomy of interest is challenging. Previous methods aimed at generating vessel trees mostly in an autoregressive style and could not be applied to vessel graphs with cycles such as capillaries or specific anatomical structures such as the Circle of Willis. Addressing this gap, we introduce the first application of \textit{denoising diffusion models} in 3D vessel graph generation. Our contributions include a novel, two-stage generation method that sequentially denoises node coordinates and edges. We experiment with two real-world vessel datasets, consisting of microscopic capillaries and major cerebral vessels, and demonstrate the generalizability of our method for producing diverse, novel, and anatomically plausible vessel graphs.
Benchmarking the CoW with the TopCoW Challenge: Topology-Aware Anatomical Segmentation of the Circle of Willis for CTA and MRA
Dec 29, 2023



Abstract:The Circle of Willis (CoW) is an important network of arteries connecting major circulations of the brain. Its vascular architecture is believed to affect the risk, severity, and clinical outcome of serious neuro-vascular diseases. However, characterizing the highly variable CoW anatomy is still a manual and time-consuming expert task. The CoW is usually imaged by two angiographic imaging modalities, magnetic resonance angiography (MRA) and computed tomography angiography (CTA), but there exist limited public datasets with annotations on CoW anatomy, especially for CTA. Therefore we organized the TopCoW Challenge in 2023 with the release of an annotated CoW dataset and invited submissions worldwide for the CoW segmentation task, which attracted over 140 registered participants from four continents. TopCoW dataset was the first public dataset with voxel-level annotations for CoW's 13 vessel components, made possible by virtual-reality (VR) technology. It was also the first dataset with paired MRA and CTA from the same patients. TopCoW challenge aimed to tackle the CoW characterization problem as a multiclass anatomical segmentation task with an emphasis on topological metrics. The top performing teams managed to segment many CoW components to Dice scores around 90%, but with lower scores for communicating arteries and rare variants. There were also topological mistakes for predictions with high Dice scores. Additional topological analysis revealed further areas for improvement in detecting certain CoW components and matching CoW variant's topology accurately. TopCoW represented a first attempt at benchmarking the CoW anatomical segmentation task for MRA and CTA, both morphologically and topologically.
DENTEX: An Abnormal Tooth Detection with Dental Enumeration and Diagnosis Benchmark for Panoramic X-rays
May 30, 2023

Abstract:Panoramic X-rays are frequently used in dentistry for treatment planning, but their interpretation can be both time-consuming and prone to error. Artificial intelligence (AI) has the potential to aid in the analysis of these X-rays, thereby improving the accuracy of dental diagnoses and treatment plans. Nevertheless, designing automated algorithms for this purpose poses significant challenges, mainly due to the scarcity of annotated data and variations in anatomical structure. To address these issues, the Dental Enumeration and Diagnosis on Panoramic X-rays Challenge (DENTEX) has been organized in association with the International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI) in 2023. This challenge aims to promote the development of algorithms for multi-label detection of abnormal teeth, using three types of hierarchically annotated data: partially annotated quadrant data, partially annotated quadrant-enumeration data, and fully annotated quadrant-enumeration-diagnosis data, inclusive of four different diagnoses. In this paper, we present the results of evaluating participant algorithms on the fully annotated data, additionally investigating performance variation for quadrant, enumeration, and diagnosis labels in the detection of abnormal teeth. The provision of this annotated dataset, alongside the results of this challenge, may lay the groundwork for the creation of AI-powered tools that can offer more precise and efficient diagnosis and treatment planning in the field of dentistry. The evaluation code and datasets can be accessed at https://github.com/ibrahimethemhamamci/DENTEX
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge