Susanne Wegener
Going beyond explainability in multi-modal stroke outcome prediction models
Apr 07, 2025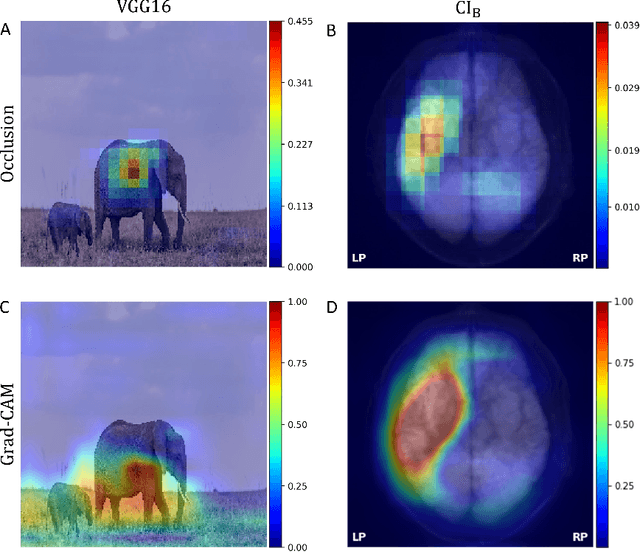
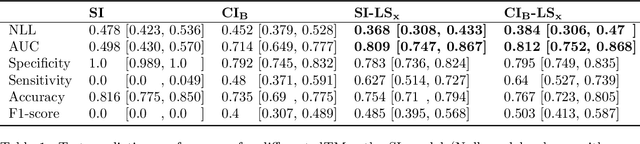
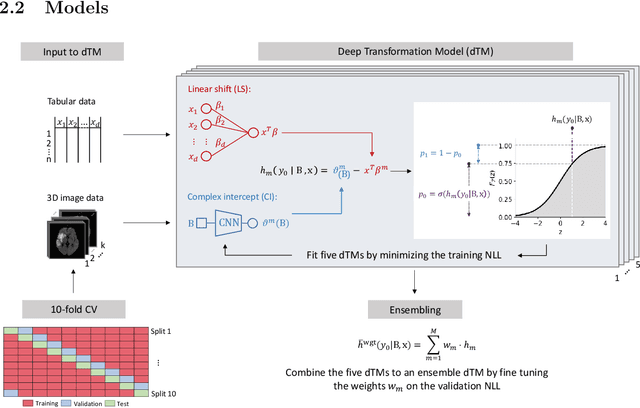
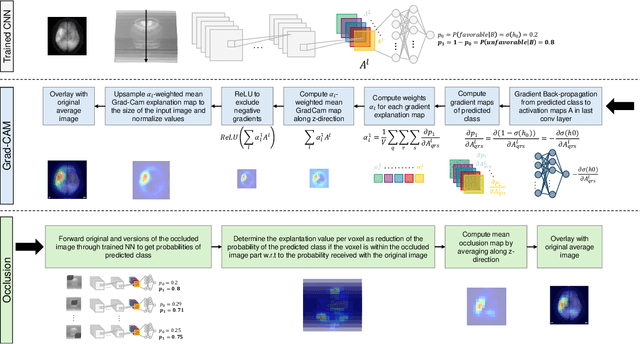
Abstract:Aim: This study aims to enhance interpretability and explainability of multi-modal prediction models integrating imaging and tabular patient data. Methods: We adapt the xAI methods Grad-CAM and Occlusion to multi-modal, partly interpretable deep transformation models (dTMs). DTMs combine statistical and deep learning approaches to simultaneously achieve state-of-the-art prediction performance and interpretable parameter estimates, such as odds ratios for tabular features. Based on brain imaging and tabular data from 407 stroke patients, we trained dTMs to predict functional outcome three months after stroke. We evaluated the models using different discriminatory metrics. The adapted xAI methods were used to generated explanation maps for identification of relevant image features and error analysis. Results: The dTMs achieve state-of-the-art prediction performance, with area under the curve (AUC) values close to 0.8. The most important tabular predictors of functional outcome are functional independence before stroke and NIHSS on admission, a neurological score indicating stroke severity. Explanation maps calculated from brain imaging dTMs for functional outcome highlighted critical brain regions such as the frontal lobe, which is known to be linked to age which in turn increases the risk for unfavorable outcomes. Similarity plots of the explanation maps revealed distinct patterns which give insight into stroke pathophysiology, support developing novel predictors of stroke outcome and enable to identify false predictions. Conclusion: By adapting methods for explanation maps to dTMs, we enhanced the explainability of multi-modal and partly interpretable prediction models. The resulting explanation maps facilitate error analysis and support hypothesis generation regarding the significance of specific image regions in outcome prediction.
ISLES'24: Improving final infarct prediction in ischemic stroke using multimodal imaging and clinical data
Aug 20, 2024
Abstract:Accurate estimation of core (irreversibly damaged tissue) and penumbra (salvageable tissue) volumes is essential for ischemic stroke treatment decisions. Perfusion CT, the clinical standard, estimates these volumes but is affected by variations in deconvolution algorithms, implementations, and thresholds. Core tissue expands over time, with growth rates influenced by thrombus location, collateral circulation, and inherent patient-specific factors. Understanding this tissue growth is crucial for determining the need to transfer patients to comprehensive stroke centers, predicting the benefits of additional reperfusion attempts during mechanical thrombectomy, and forecasting final clinical outcomes. This work presents the ISLES'24 challenge, which addresses final post-treatment stroke infarct prediction from pre-interventional acute stroke imaging and clinical data. ISLES'24 establishes a unique 360-degree setting where all feasibly accessible clinical data are available for participants, including full CT acute stroke imaging, sub-acute follow-up MRI, and clinical tabular data. The contributions of this work are two-fold: first, we introduce a standardized benchmarking of final stroke infarct segmentation algorithms through the ISLES'24 challenge; second, we provide insights into infarct segmentation using multimodal imaging and clinical data strategies by identifying outperforming methods on a finely curated dataset. The outputs of this challenge are anticipated to enhance clinical decision-making and improve patient outcome predictions. All ISLES'24 materials, including data, performance evaluation scripts, and leading algorithmic strategies, are available to the research community following \url{https://isles-24.grand-challenge.org/}.
ISLES 2024: The first longitudinal multimodal multi-center real-world dataset in (sub-)acute stroke
Aug 20, 2024
Abstract:Stroke remains a leading cause of global morbidity and mortality, placing a heavy socioeconomic burden. Over the past decade, advances in endovascular reperfusion therapy and the use of CT and MRI imaging for treatment guidance have significantly improved patient outcomes and are now standard in clinical practice. To develop machine learning algorithms that can extract meaningful and reproducible models of brain function for both clinical and research purposes from stroke images - particularly for lesion identification, brain health quantification, and prognosis - large, diverse, and well-annotated public datasets are essential. While only a few datasets with (sub-)acute stroke data were previously available, several large, high-quality datasets have recently been made publicly accessible. However, these existing datasets include only MRI data. In contrast, our dataset is the first to offer comprehensive longitudinal stroke data, including acute CT imaging with angiography and perfusion, follow-up MRI at 2-9 days, as well as acute and longitudinal clinical data up to a three-month outcome. The dataset includes a training dataset of n = 150 and a test dataset of n = 100 scans. Training data is publicly available, while test data will be used exclusively for model validation. We are making this dataset available as part of the 2024 edition of the Ischemic Stroke Lesion Segmentation (ISLES) challenge (https://www.isles-challenge.org/), which continuously aims to establish benchmark methods for acute and sub-acute ischemic stroke lesion segmentation, aiding in creating open stroke imaging datasets and evaluating cutting-edge image processing algorithms.
A Robust Ensemble Algorithm for Ischemic Stroke Lesion Segmentation: Generalizability and Clinical Utility Beyond the ISLES Challenge
Apr 03, 2024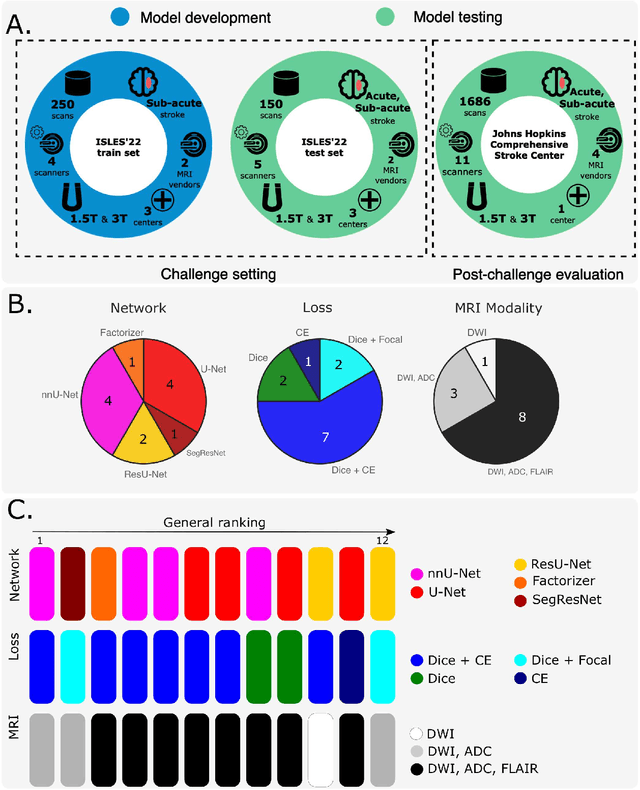
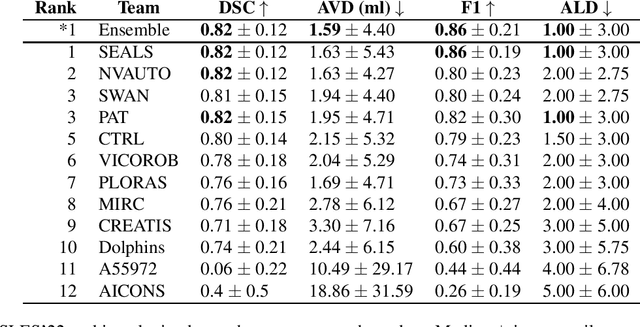
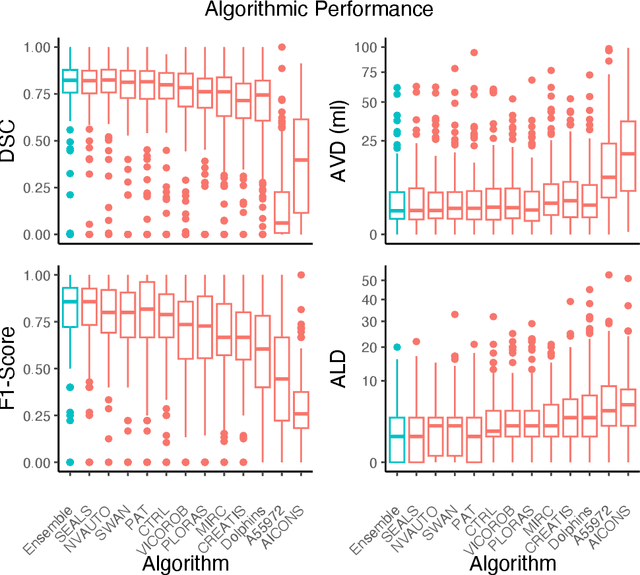
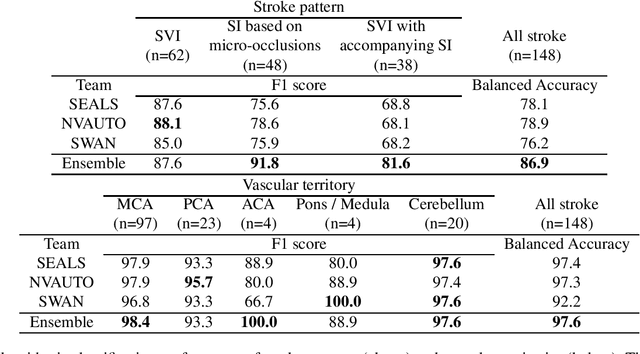
Abstract:Diffusion-weighted MRI (DWI) is essential for stroke diagnosis, treatment decisions, and prognosis. However, image and disease variability hinder the development of generalizable AI algorithms with clinical value. We address this gap by presenting a novel ensemble algorithm derived from the 2022 Ischemic Stroke Lesion Segmentation (ISLES) challenge. ISLES'22 provided 400 patient scans with ischemic stroke from various medical centers, facilitating the development of a wide range of cutting-edge segmentation algorithms by the research community. Through collaboration with leading teams, we combined top-performing algorithms into an ensemble model that overcomes the limitations of individual solutions. Our ensemble model achieved superior ischemic lesion detection and segmentation accuracy on our internal test set compared to individual algorithms. This accuracy generalized well across diverse image and disease variables. Furthermore, the model excelled in extracting clinical biomarkers. Notably, in a Turing-like test, neuroradiologists consistently preferred the algorithm's segmentations over manual expert efforts, highlighting increased comprehensiveness and precision. Validation using a real-world external dataset (N=1686) confirmed the model's generalizability. The algorithm's outputs also demonstrated strong correlations with clinical scores (admission NIHSS and 90-day mRS) on par with or exceeding expert-derived results, underlining its clinical relevance. This study offers two key findings. First, we present an ensemble algorithm (https://github.com/Tabrisrei/ISLES22_Ensemble) that detects and segments ischemic stroke lesions on DWI across diverse scenarios on par with expert (neuro)radiologists. Second, we show the potential for biomedical challenge outputs to extend beyond the challenge's initial objectives, demonstrating their real-world clinical applicability.
Benchmarking the CoW with the TopCoW Challenge: Topology-Aware Anatomical Segmentation of the Circle of Willis for CTA and MRA
Dec 29, 2023



Abstract:The Circle of Willis (CoW) is an important network of arteries connecting major circulations of the brain. Its vascular architecture is believed to affect the risk, severity, and clinical outcome of serious neuro-vascular diseases. However, characterizing the highly variable CoW anatomy is still a manual and time-consuming expert task. The CoW is usually imaged by two angiographic imaging modalities, magnetic resonance angiography (MRA) and computed tomography angiography (CTA), but there exist limited public datasets with annotations on CoW anatomy, especially for CTA. Therefore we organized the TopCoW Challenge in 2023 with the release of an annotated CoW dataset and invited submissions worldwide for the CoW segmentation task, which attracted over 140 registered participants from four continents. TopCoW dataset was the first public dataset with voxel-level annotations for CoW's 13 vessel components, made possible by virtual-reality (VR) technology. It was also the first dataset with paired MRA and CTA from the same patients. TopCoW challenge aimed to tackle the CoW characterization problem as a multiclass anatomical segmentation task with an emphasis on topological metrics. The top performing teams managed to segment many CoW components to Dice scores around 90%, but with lower scores for communicating arteries and rare variants. There were also topological mistakes for predictions with high Dice scores. Additional topological analysis revealed further areas for improvement in detecting certain CoW components and matching CoW variant's topology accurately. TopCoW represented a first attempt at benchmarking the CoW anatomical segmentation task for MRA and CTA, both morphologically and topologically.
Integrating uncertainty in deep neural networks for MRI based stroke analysis
Aug 13, 2020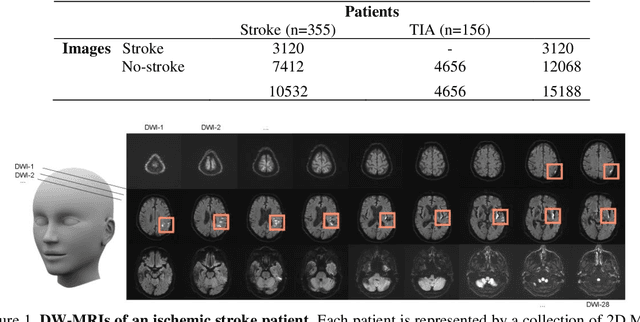
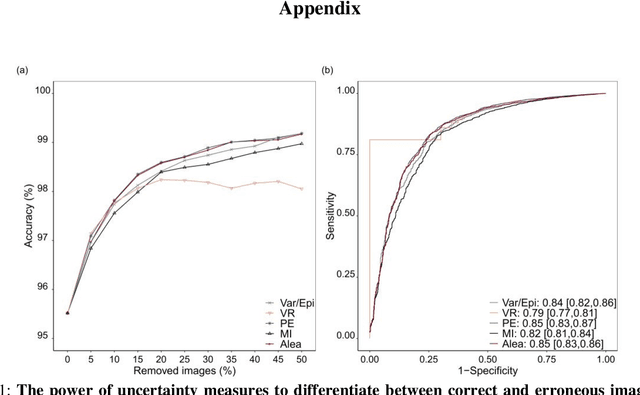
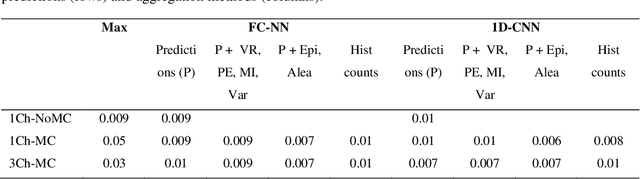
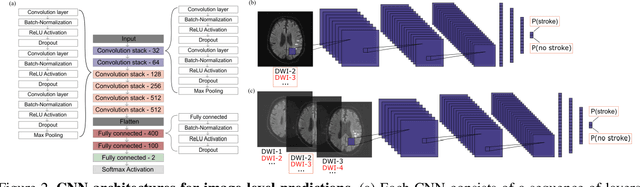
Abstract:At present, the majority of the proposed Deep Learning (DL) methods provide point predictions without quantifying the models uncertainty. However, a quantification of the reliability of automated image analysis is essential, in particular in medicine when physicians rely on the results for making critical treatment decisions. In this work, we provide an entire framework to diagnose ischemic stroke patients incorporating Bayesian uncertainty into the analysis procedure. We present a Bayesian Convolutional Neural Network (CNN) yielding a probability for a stroke lesion on 2D Magnetic Resonance (MR) images with corresponding uncertainty information about the reliability of the prediction. For patient-level diagnoses, different aggregation methods are proposed and evaluated, which combine the single image-level predictions. Those methods take advantage of the uncertainty in image predictions and report model uncertainty at the patient-level. In a cohort of 511 patients, our Bayesian CNN achieved an accuracy of 95.33% at the image-level representing a significant improvement of 2% over a non-Bayesian counterpart. The best patient aggregation method yielded 95.89% of accuracy. Integrating uncertainty information about image predictions in aggregation models resulted in higher uncertainty measures to false patient classifications, which enabled to filter critical patient diagnoses that are supposed to be closer examined by a medical doctor. We therefore recommend using Bayesian approaches not only for improved image-level prediction and uncertainty estimation but also for the detection of uncertain aggregations at the patient-level.
* 21 pages, 13 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge