Lisa Herzog
Going beyond explainability in multi-modal stroke outcome prediction models
Apr 07, 2025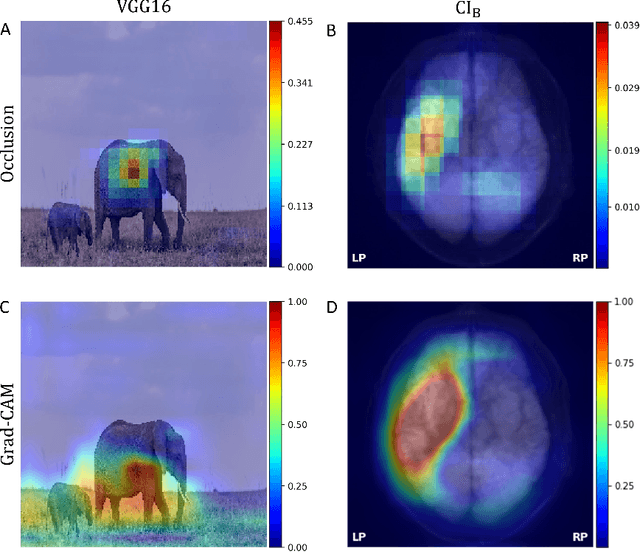
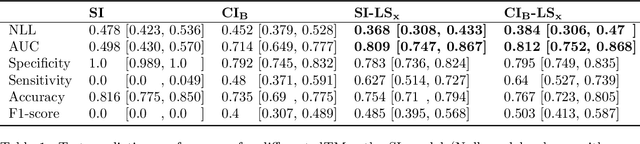
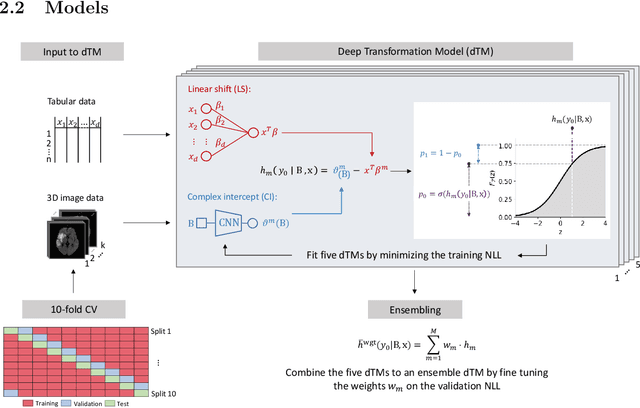
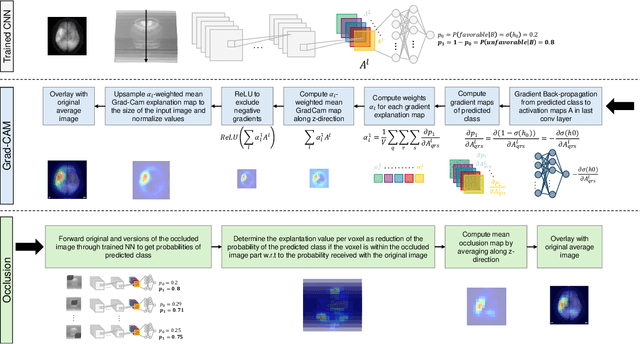
Abstract:Aim: This study aims to enhance interpretability and explainability of multi-modal prediction models integrating imaging and tabular patient data. Methods: We adapt the xAI methods Grad-CAM and Occlusion to multi-modal, partly interpretable deep transformation models (dTMs). DTMs combine statistical and deep learning approaches to simultaneously achieve state-of-the-art prediction performance and interpretable parameter estimates, such as odds ratios for tabular features. Based on brain imaging and tabular data from 407 stroke patients, we trained dTMs to predict functional outcome three months after stroke. We evaluated the models using different discriminatory metrics. The adapted xAI methods were used to generated explanation maps for identification of relevant image features and error analysis. Results: The dTMs achieve state-of-the-art prediction performance, with area under the curve (AUC) values close to 0.8. The most important tabular predictors of functional outcome are functional independence before stroke and NIHSS on admission, a neurological score indicating stroke severity. Explanation maps calculated from brain imaging dTMs for functional outcome highlighted critical brain regions such as the frontal lobe, which is known to be linked to age which in turn increases the risk for unfavorable outcomes. Similarity plots of the explanation maps revealed distinct patterns which give insight into stroke pathophysiology, support developing novel predictors of stroke outcome and enable to identify false predictions. Conclusion: By adapting methods for explanation maps to dTMs, we enhanced the explainability of multi-modal and partly interpretable prediction models. The resulting explanation maps facilitate error analysis and support hypothesis generation regarding the significance of specific image regions in outcome prediction.
Ordinal Neural Network Transformation Models: Deep and interpretable regression models for ordinal outcomes
Oct 26, 2020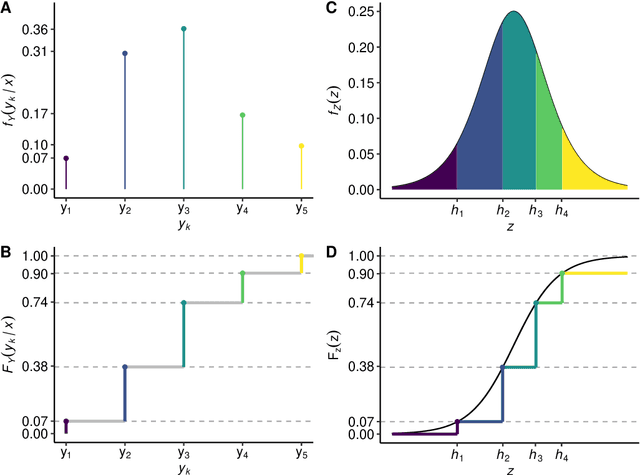
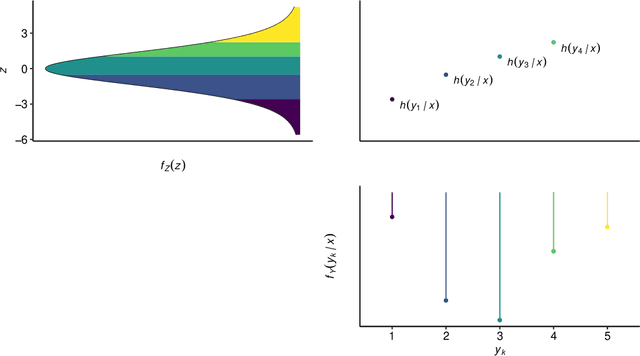
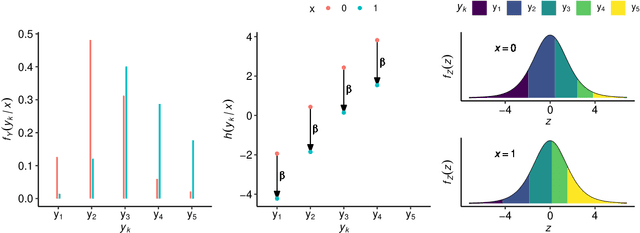
Abstract:Outcomes with a natural order commonly occur in prediction tasks and oftentimes the available input data are a mixture of complex data, like images, and tabular predictors. Although deep Learning (DL) methods have shown outstanding performance on image classification, most models treat ordered outcomes as unordered and lack interpretability. In contrast, classical ordinal regression models yield interpretable predictor effects but are limited to tabular input data. Here, we present the highly modular class of ordinal neural network transformation models (ONTRAMs). Transformation models use a parametric transformation function and a simple distribution to trade off flexibility and interpretability of individual model components. In ONTRAMs, this trade-off is achieved by additively decomposing the transformation function into terms for the tabular and image data using a set of jointly trained neural networks. We show that the most flexible ONTRAMs achieve on-par performance with DL classifiers while outperforming them in training speed. We discuss how to interpret components of ONTRAMs in general and in the case of correlated tabular and image data. Taken together, ONTRAMs join benefits of DL and distributional regression to create interpretable prediction models for ordinal outcomes.
Integrating uncertainty in deep neural networks for MRI based stroke analysis
Aug 13, 2020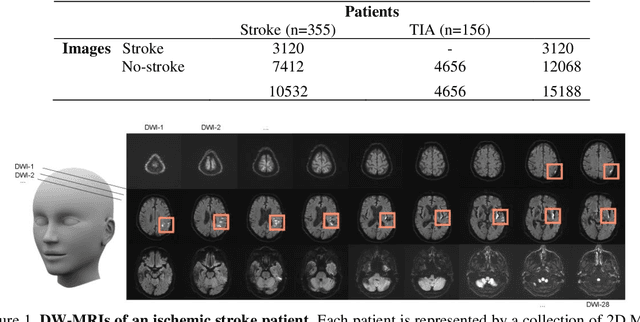
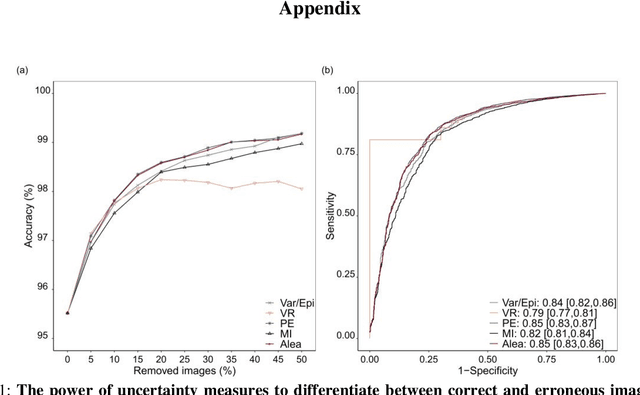
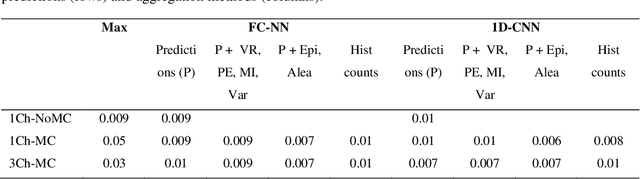
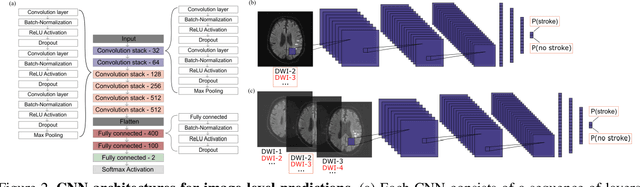
Abstract:At present, the majority of the proposed Deep Learning (DL) methods provide point predictions without quantifying the models uncertainty. However, a quantification of the reliability of automated image analysis is essential, in particular in medicine when physicians rely on the results for making critical treatment decisions. In this work, we provide an entire framework to diagnose ischemic stroke patients incorporating Bayesian uncertainty into the analysis procedure. We present a Bayesian Convolutional Neural Network (CNN) yielding a probability for a stroke lesion on 2D Magnetic Resonance (MR) images with corresponding uncertainty information about the reliability of the prediction. For patient-level diagnoses, different aggregation methods are proposed and evaluated, which combine the single image-level predictions. Those methods take advantage of the uncertainty in image predictions and report model uncertainty at the patient-level. In a cohort of 511 patients, our Bayesian CNN achieved an accuracy of 95.33% at the image-level representing a significant improvement of 2% over a non-Bayesian counterpart. The best patient aggregation method yielded 95.89% of accuracy. Integrating uncertainty information about image predictions in aggregation models resulted in higher uncertainty measures to false patient classifications, which enabled to filter critical patient diagnoses that are supposed to be closer examined by a medical doctor. We therefore recommend using Bayesian approaches not only for improved image-level prediction and uncertainty estimation but also for the detection of uncertain aggregations at the patient-level.
* 21 pages, 13 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge