David Robben
ISLES'24: Improving final infarct prediction in ischemic stroke using multimodal imaging and clinical data
Aug 20, 2024
Abstract:Accurate estimation of core (irreversibly damaged tissue) and penumbra (salvageable tissue) volumes is essential for ischemic stroke treatment decisions. Perfusion CT, the clinical standard, estimates these volumes but is affected by variations in deconvolution algorithms, implementations, and thresholds. Core tissue expands over time, with growth rates influenced by thrombus location, collateral circulation, and inherent patient-specific factors. Understanding this tissue growth is crucial for determining the need to transfer patients to comprehensive stroke centers, predicting the benefits of additional reperfusion attempts during mechanical thrombectomy, and forecasting final clinical outcomes. This work presents the ISLES'24 challenge, which addresses final post-treatment stroke infarct prediction from pre-interventional acute stroke imaging and clinical data. ISLES'24 establishes a unique 360-degree setting where all feasibly accessible clinical data are available for participants, including full CT acute stroke imaging, sub-acute follow-up MRI, and clinical tabular data. The contributions of this work are two-fold: first, we introduce a standardized benchmarking of final stroke infarct segmentation algorithms through the ISLES'24 challenge; second, we provide insights into infarct segmentation using multimodal imaging and clinical data strategies by identifying outperforming methods on a finely curated dataset. The outputs of this challenge are anticipated to enhance clinical decision-making and improve patient outcome predictions. All ISLES'24 materials, including data, performance evaluation scripts, and leading algorithmic strategies, are available to the research community following \url{https://isles-24.grand-challenge.org/}.
ISLES 2024: The first longitudinal multimodal multi-center real-world dataset in (sub-)acute stroke
Aug 20, 2024
Abstract:Stroke remains a leading cause of global morbidity and mortality, placing a heavy socioeconomic burden. Over the past decade, advances in endovascular reperfusion therapy and the use of CT and MRI imaging for treatment guidance have significantly improved patient outcomes and are now standard in clinical practice. To develop machine learning algorithms that can extract meaningful and reproducible models of brain function for both clinical and research purposes from stroke images - particularly for lesion identification, brain health quantification, and prognosis - large, diverse, and well-annotated public datasets are essential. While only a few datasets with (sub-)acute stroke data were previously available, several large, high-quality datasets have recently been made publicly accessible. However, these existing datasets include only MRI data. In contrast, our dataset is the first to offer comprehensive longitudinal stroke data, including acute CT imaging with angiography and perfusion, follow-up MRI at 2-9 days, as well as acute and longitudinal clinical data up to a three-month outcome. The dataset includes a training dataset of n = 150 and a test dataset of n = 100 scans. Training data is publicly available, while test data will be used exclusively for model validation. We are making this dataset available as part of the 2024 edition of the Ischemic Stroke Lesion Segmentation (ISLES) challenge (https://www.isles-challenge.org/), which continuously aims to establish benchmark methods for acute and sub-acute ischemic stroke lesion segmentation, aiding in creating open stroke imaging datasets and evaluating cutting-edge image processing algorithms.
A Robust Ensemble Algorithm for Ischemic Stroke Lesion Segmentation: Generalizability and Clinical Utility Beyond the ISLES Challenge
Apr 03, 2024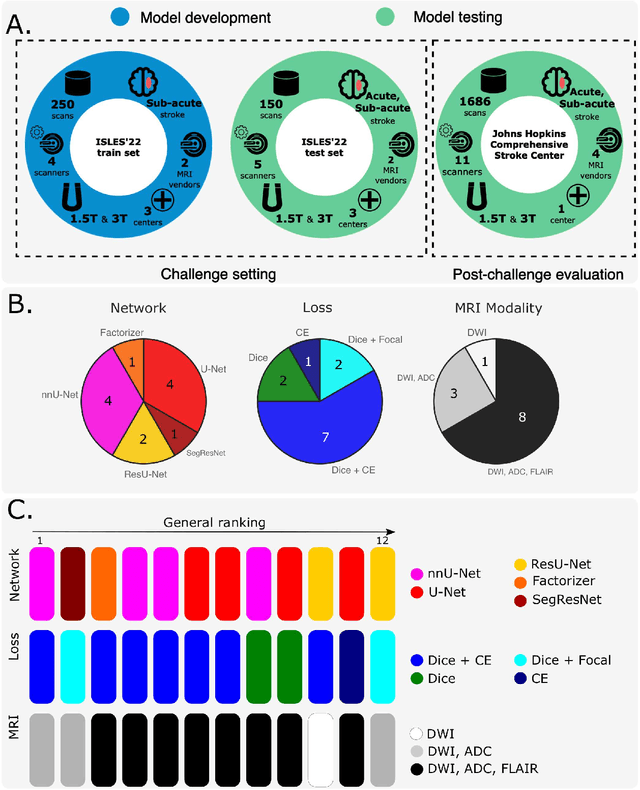
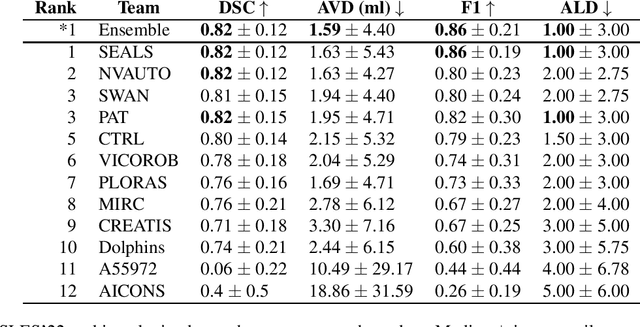
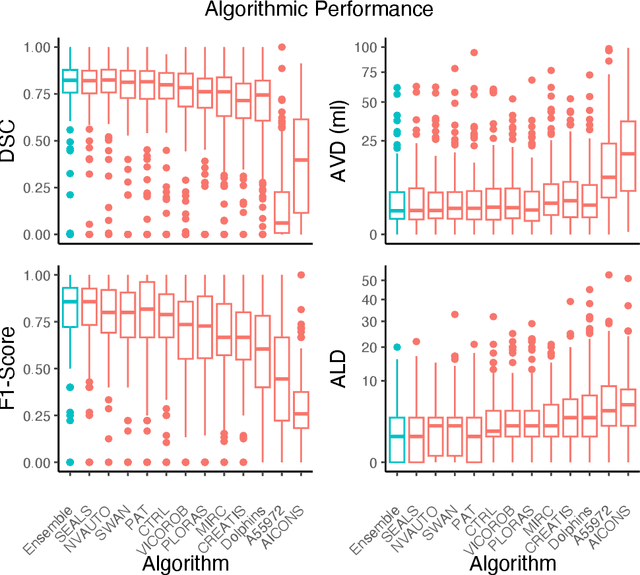
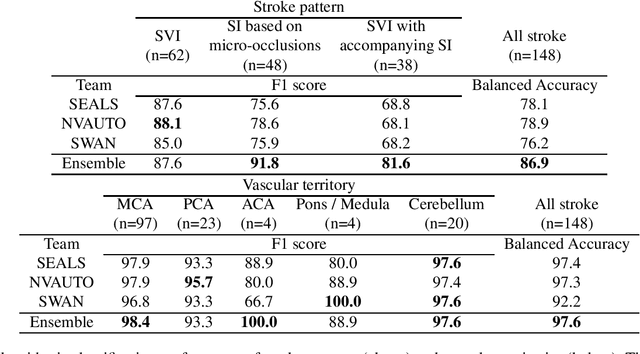
Abstract:Diffusion-weighted MRI (DWI) is essential for stroke diagnosis, treatment decisions, and prognosis. However, image and disease variability hinder the development of generalizable AI algorithms with clinical value. We address this gap by presenting a novel ensemble algorithm derived from the 2022 Ischemic Stroke Lesion Segmentation (ISLES) challenge. ISLES'22 provided 400 patient scans with ischemic stroke from various medical centers, facilitating the development of a wide range of cutting-edge segmentation algorithms by the research community. Through collaboration with leading teams, we combined top-performing algorithms into an ensemble model that overcomes the limitations of individual solutions. Our ensemble model achieved superior ischemic lesion detection and segmentation accuracy on our internal test set compared to individual algorithms. This accuracy generalized well across diverse image and disease variables. Furthermore, the model excelled in extracting clinical biomarkers. Notably, in a Turing-like test, neuroradiologists consistently preferred the algorithm's segmentations over manual expert efforts, highlighting increased comprehensiveness and precision. Validation using a real-world external dataset (N=1686) confirmed the model's generalizability. The algorithm's outputs also demonstrated strong correlations with clinical scores (admission NIHSS and 90-day mRS) on par with or exceeding expert-derived results, underlining its clinical relevance. This study offers two key findings. First, we present an ensemble algorithm (https://github.com/Tabrisrei/ISLES22_Ensemble) that detects and segments ischemic stroke lesions on DWI across diverse scenarios on par with expert (neuro)radiologists. Second, we show the potential for biomedical challenge outputs to extend beyond the challenge's initial objectives, demonstrating their real-world clinical applicability.
Convolutional neural networks for medical image segmentation
Nov 17, 2022



Abstract:In this article, we look into some essential aspects of convolutional neural networks (CNNs) with the focus on medical image segmentation. First, we discuss the CNN architecture, thereby highlighting the spatial origin of the data, voxel-wise classification and the receptive field. Second, we discuss the sampling of input-output pairs, thereby highlighting the interaction between voxel-wise classification, patch size and the receptive field. Finally, we give a historical overview of crucial changes to CNN architectures for classification and segmentation, giving insights in the relation between three pivotal CNN architectures: FCN, U-Net and DeepMedic.
DeepVoxNet2: Yet another CNN framework
Nov 17, 2022Abstract:We know that both the CNN mapping function and the sampling scheme are of paramount importance for CNN-based image analysis. It is clear that both functions operate in the same space, with an image axis $\mathcal{I}$ and a feature axis $\mathcal{F}$. Remarkably, we found that no frameworks existed that unified the two and kept track of the spatial origin of the data automatically. Based on our own practical experience, we found the latter to often result in complex coding and pipelines that are difficult to exchange. This article introduces our framework for 1, 2 or 3D image classification or segmentation: DeepVoxNet2 (DVN2). This article serves as an interactive tutorial, and a pre-compiled version, including the outputs of the code blocks, can be found online in the public DVN2 repository. This tutorial uses data from the multimodal Brain Tumor Image Segmentation Benchmark (BRATS) of 2018 to show an example of a 3D segmentation pipeline.
Final infarct prediction in acute ischemic stroke
Nov 09, 2022Abstract:This article focuses on the control center of each human body: the brain. We will point out the pivotal role of the cerebral vasculature and how its complex mechanisms may vary between subjects. We then emphasize a specific acute pathological state, i.e., acute ischemic stroke, and show how medical imaging and its analysis can be used to define the treatment. We show how the core-penumbra concept is used in practice using mismatch criteria and how machine learning can be used to make predictions of the final infarct, either via deconvolution or convolutional neural networks.
Theoretical analysis and experimental validation of volume bias of soft Dice optimized segmentation maps in the context of inherent uncertainty
Nov 08, 2022



Abstract:The clinical interest is often to measure the volume of a structure, which is typically derived from a segmentation. In order to evaluate and compare segmentation methods, the similarity between a segmentation and a predefined ground truth is measured using popular discrete metrics, such as the Dice score. Recent segmentation methods use a differentiable surrogate metric, such as soft Dice, as part of the loss function during the learning phase. In this work, we first briefly describe how to derive volume estimates from a segmentation that is, potentially, inherently uncertain or ambiguous. This is followed by a theoretical analysis and an experimental validation linking the inherent uncertainty to common loss functions for training CNNs, namely cross-entropy and soft Dice. We find that, even though soft Dice optimization leads to an improved performance with respect to the Dice score and other measures, it may introduce a volume bias for tasks with high inherent uncertainty. These findings indicate some of the method's clinical limitations and suggest doing a closer ad-hoc volume analysis with an optional re-calibration step.
* 18 pages, 7 figures, 3 tables, published in Elsevier Medical Image Analysis (2021)
The Dice loss in the context of missing or empty labels: Introducing $Φ$ and $ε$
Jul 19, 2022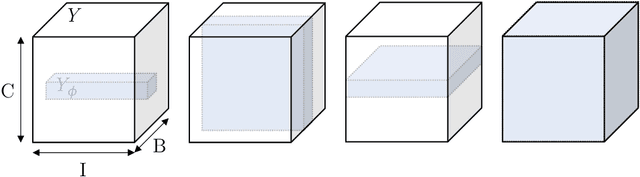
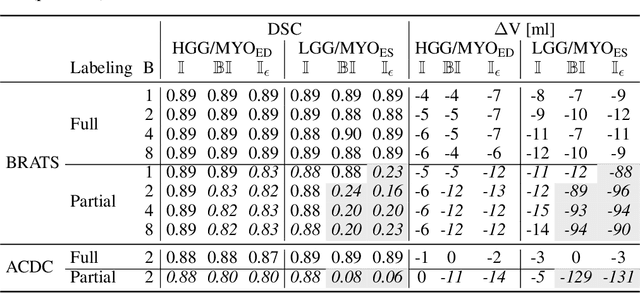
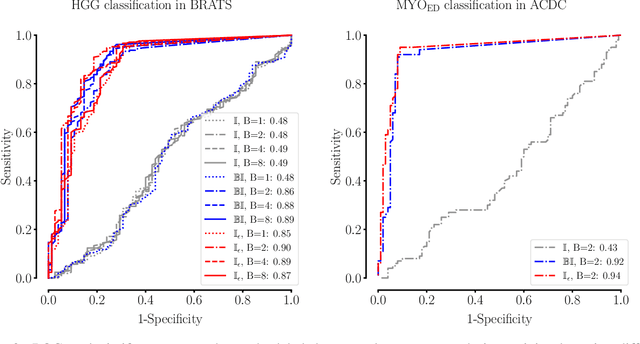
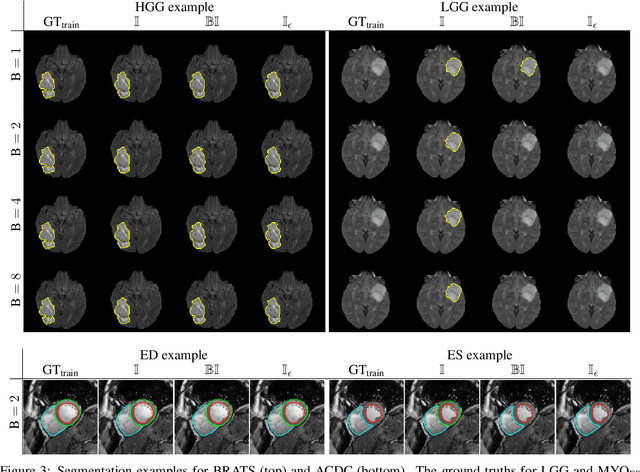
Abstract:Albeit the Dice loss is one of the dominant loss functions in medical image segmentation, most research omits a closer look at its derivative, i.e. the real motor of the optimization when using gradient descent. In this paper, we highlight the peculiar action of the Dice loss in the presence of missing or empty labels. First, we formulate a theoretical basis that gives a general description of the Dice loss and its derivative. It turns out that the choice of the reduction dimensions $\Phi$ and the smoothing term $\epsilon$ is non-trivial and greatly influences its behavior. We find and propose heuristic combinations of $\Phi$ and $\epsilon$ that work in a segmentation setting with either missing or empty labels. Second, we empirically validate these findings in a binary and multiclass segmentation setting using two publicly available datasets. We confirm that the choice of $\Phi$ and $\epsilon$ is indeed pivotal. With $\Phi$ chosen such that the reductions happen over a single batch (and class) element and with a negligible $\epsilon$, the Dice loss deals with missing labels naturally and performs similarly compared to recent adaptations specific for missing labels. With $\Phi$ chosen such that the reductions happen over multiple batch elements or with a heuristic value for $\epsilon$, the Dice loss handles empty labels correctly. We believe that this work highlights some essential perspectives and hope that it encourages researchers to better describe their exact implementation of the Dice loss in future work.
ISLES 2022: A multi-center magnetic resonance imaging stroke lesion segmentation dataset
Jun 14, 2022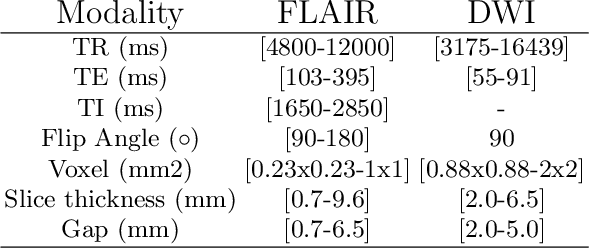
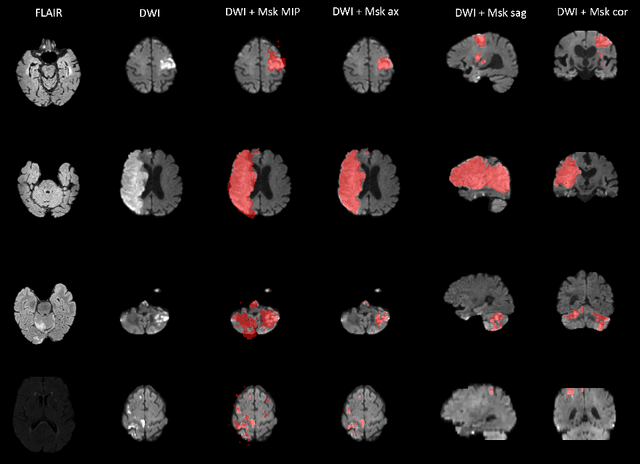

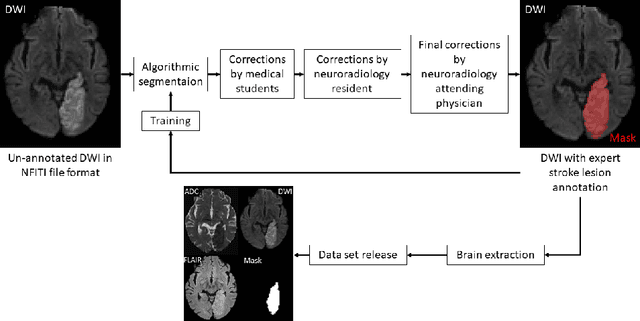
Abstract:Magnetic resonance imaging (MRI) is a central modality for stroke imaging. It is used upon patient admission to make treatment decisions such as selecting patients for intravenous thrombolysis or endovascular therapy. MRI is later used in the duration of hospital stay to predict outcome by visualizing infarct core size and location. Furthermore, it may be used to characterize stroke etiology, e.g. differentiation between (cardio)-embolic and non-embolic stroke. Computer based automated medical image processing is increasingly finding its way into clinical routine. Previous iterations of the Ischemic Stroke Lesion Segmentation (ISLES) challenge have aided in the generation of identifying benchmark methods for acute and sub-acute ischemic stroke lesion segmentation. Here we introduce an expert-annotated, multicenter MRI dataset for segmentation of acute to subacute stroke lesions. This dataset comprises 400 multi-vendor MRI cases with high variability in stroke lesion size, quantity and location. It is split into a training dataset of n=250 and a test dataset of n=150. All training data will be made publicly available. The test dataset will be used for model validation only and will not be released to the public. This dataset serves as the foundation of the ISLES 2022 challenge with the goal of finding algorithmic methods to enable the development and benchmarking of robust and accurate segmentation algorithms for ischemic stroke.
Differentiable Deconvolution for Improved Stroke Perfusion Analysis
Mar 31, 2021



Abstract:Perfusion imaging is the current gold standard for acute ischemic stroke analysis. It allows quantification of the salvageable and non-salvageable tissue regions (penumbra and core areas respectively). In clinical settings, the singular value decomposition (SVD) deconvolution is one of the most accepted and used approaches for generating interpretable and physically meaningful maps. Though this method has been widely validated in experimental and clinical settings, it might produce suboptimal results because the chosen inputs to the model cannot guarantee optimal performance. For the most critical input, the arterial input function (AIF), it is still controversial how and where it should be chosen even though the method is very sensitive to this input. In this work we propose an AIF selection approach that is optimized for maximal core lesion segmentation performance. The AIF is regressed by a neural network optimized through a differentiable SVD deconvolution, aiming to maximize core lesion segmentation agreement with ground truth data. To our knowledge, this is the first work exploiting a differentiable deconvolution model with neural networks. We show that our approach is able to generate AIFs without any manual annotation, and hence avoiding manual rater's influences. The method achieves manual expert performance in the ISLES18 dataset. We conclude that the methodology opens new possibilities for improving perfusion imaging quantification with deep neural networks.
* Accepted at MICCAI 2020
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge