Diana M. Sima
A Robust Ensemble Algorithm for Ischemic Stroke Lesion Segmentation: Generalizability and Clinical Utility Beyond the ISLES Challenge
Apr 03, 2024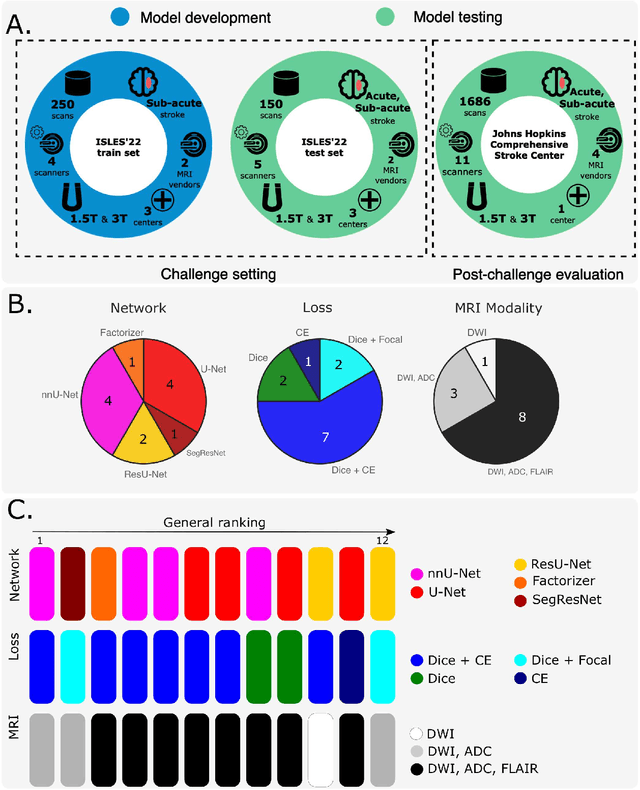
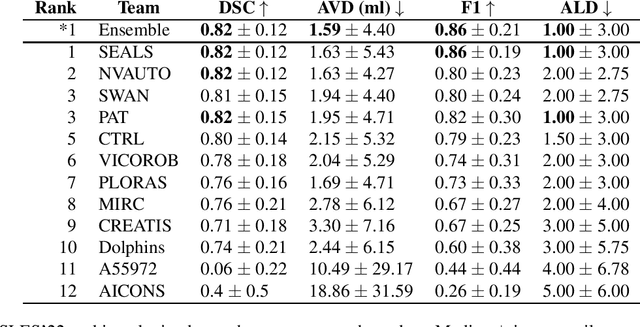
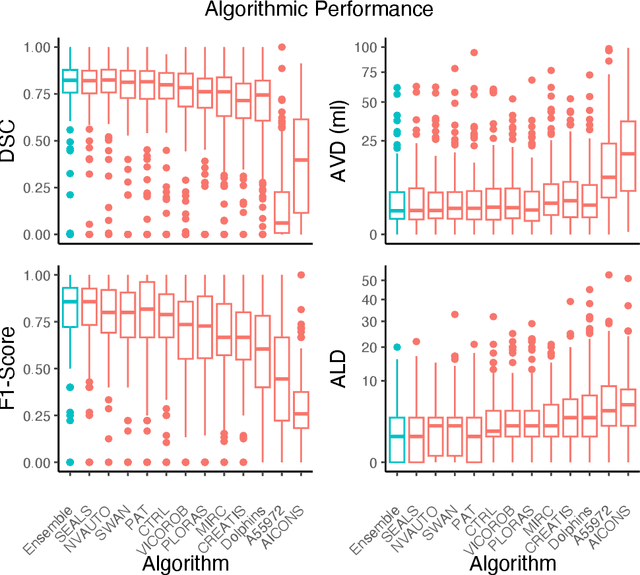
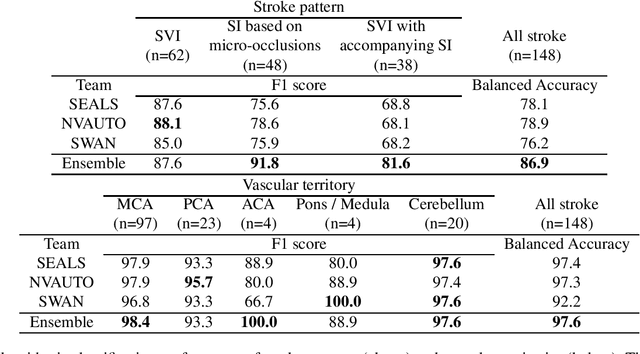
Abstract:Diffusion-weighted MRI (DWI) is essential for stroke diagnosis, treatment decisions, and prognosis. However, image and disease variability hinder the development of generalizable AI algorithms with clinical value. We address this gap by presenting a novel ensemble algorithm derived from the 2022 Ischemic Stroke Lesion Segmentation (ISLES) challenge. ISLES'22 provided 400 patient scans with ischemic stroke from various medical centers, facilitating the development of a wide range of cutting-edge segmentation algorithms by the research community. Through collaboration with leading teams, we combined top-performing algorithms into an ensemble model that overcomes the limitations of individual solutions. Our ensemble model achieved superior ischemic lesion detection and segmentation accuracy on our internal test set compared to individual algorithms. This accuracy generalized well across diverse image and disease variables. Furthermore, the model excelled in extracting clinical biomarkers. Notably, in a Turing-like test, neuroradiologists consistently preferred the algorithm's segmentations over manual expert efforts, highlighting increased comprehensiveness and precision. Validation using a real-world external dataset (N=1686) confirmed the model's generalizability. The algorithm's outputs also demonstrated strong correlations with clinical scores (admission NIHSS and 90-day mRS) on par with or exceeding expert-derived results, underlining its clinical relevance. This study offers two key findings. First, we present an ensemble algorithm (https://github.com/Tabrisrei/ISLES22_Ensemble) that detects and segments ischemic stroke lesions on DWI across diverse scenarios on par with expert (neuro)radiologists. Second, we show the potential for biomedical challenge outputs to extend beyond the challenge's initial objectives, demonstrating their real-world clinical applicability.
Differentiable Deconvolution for Improved Stroke Perfusion Analysis
Mar 31, 2021



Abstract:Perfusion imaging is the current gold standard for acute ischemic stroke analysis. It allows quantification of the salvageable and non-salvageable tissue regions (penumbra and core areas respectively). In clinical settings, the singular value decomposition (SVD) deconvolution is one of the most accepted and used approaches for generating interpretable and physically meaningful maps. Though this method has been widely validated in experimental and clinical settings, it might produce suboptimal results because the chosen inputs to the model cannot guarantee optimal performance. For the most critical input, the arterial input function (AIF), it is still controversial how and where it should be chosen even though the method is very sensitive to this input. In this work we propose an AIF selection approach that is optimized for maximal core lesion segmentation performance. The AIF is regressed by a neural network optimized through a differentiable SVD deconvolution, aiming to maximize core lesion segmentation agreement with ground truth data. To our knowledge, this is the first work exploiting a differentiable deconvolution model with neural networks. We show that our approach is able to generate AIFs without any manual annotation, and hence avoiding manual rater's influences. The method achieves manual expert performance in the ISLES18 dataset. We conclude that the methodology opens new possibilities for improving perfusion imaging quantification with deep neural networks.
* Accepted at MICCAI 2020
An augmentation strategy to mimic multi-scanner variability in MRI
Mar 23, 2021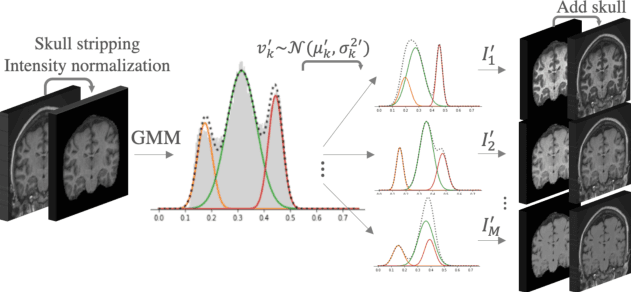
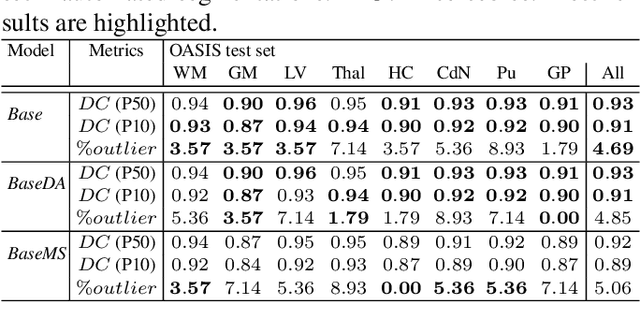
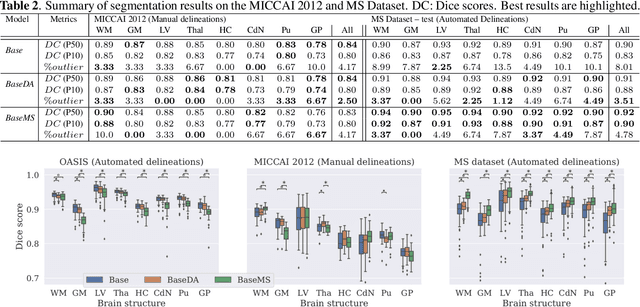
Abstract:Most publicly available brain MRI datasets are very homogeneous in terms of scanner and protocols, and it is difficult for models that learn from such data to generalize to multi-center and multi-scanner data. We propose a novel data augmentation approach with the aim of approximating the variability in terms of intensities and contrasts present in real world clinical data. We use a Gaussian Mixture Model based approach to change tissue intensities individually, producing new contrasts while preserving anatomical information. We train a deep learning model on a single scanner dataset and evaluate it on a multi-center and multi-scanner dataset. The proposed approach improves the generalization capability of the model to other scanners not present in the training data.
Unsupervised 3D Brain Anomaly Detection
Oct 09, 2020



Abstract:Anomaly detection (AD) is the identification of data samples that do not fit a learned data distribution. As such, AD systems can help physicians to determine the presence, severity, and extension of a pathology. Deep generative models, such as Generative Adversarial Networks (GANs), can be exploited to capture anatomical variability. Consequently, any outlier (i.e., sample falling outside of the learned distribution) can be detected as an abnormality in an unsupervised fashion. By using this method, we can not only detect expected or known lesions, but we can even unveil previously unrecognized biomarkers. To the best of our knowledge, this study exemplifies the first AD approach that can efficiently handle volumetric data and detect 3D brain anomalies in one single model. Our proposal is a volumetric and high-detail extension of the 2D f-AnoGAN model obtained by combining a state-of-the-art 3D GAN with refinement training steps. In experiments using non-contrast computed tomography images from traumatic brain injury (TBI) patients, the model detects and localizes TBI abnormalities with an area under the ROC curve of ~75%. Moreover, we test the potential of the method for detecting other anomalies such as low quality images, preprocessing inaccuracies, artifacts, and even the presence of post-operative signs (such as a craniectomy or a brain shunt). The method has potential for rapidly labeling abnormalities in massive imaging datasets, as well as identifying new biomarkers.
AIFNet: Automatic Vascular Function Estimation for Perfusion Analysis Using Deep Learning
Oct 04, 2020



Abstract:Perfusion imaging is crucial in acute ischemic stroke for quantifying the salvageable penumbra and irreversibly damaged core lesions. As such, it helps clinicians to decide on the optimal reperfusion treatment. In perfusion CT imaging, deconvolution methods are used to obtain clinically interpretable perfusion parameters that allow identifying brain tissue abnormalities. Deconvolution methods require the selection of two reference vascular functions as inputs to the model: the arterial input function (AIF) and the venous output function, with the AIF as the most critical model input. When manually performed, the vascular function selection is time demanding, suffers from poor reproducibility and is subject to the professionals' experience. This leads to potentially unreliable quantification of the penumbra and core lesions and, hence, might harm the treatment decision process. In this work we automatize the perfusion analysis with AIFNet, a fully automatic and end-to-end trainable deep learning approach for estimating the vascular functions. Unlike previous methods using clustering or segmentation techniques to select vascular voxels, AIFNet is directly optimized at the vascular function estimation, which allows to better recognise the time-curve profiles. Validation on the public ISLES18 stroke database shows that AIFNet reaches inter-rater performance for the vascular function estimation and, subsequently, for the parameter maps and core lesion quantification obtained through deconvolution. We conclude that AIFNet has potential for clinical transfer and could be incorporated in perfusion deconvolution software.
Improved inter-scanner MS lesion segmentation by adversarial training on longitudinal data
Feb 03, 2020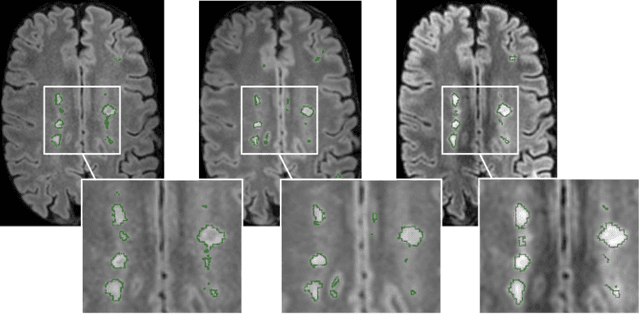
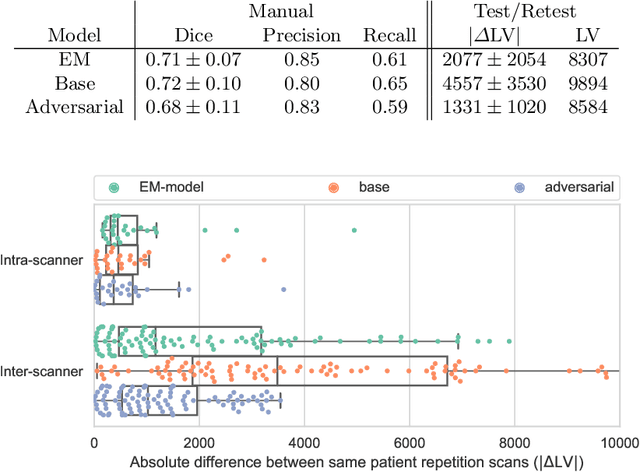
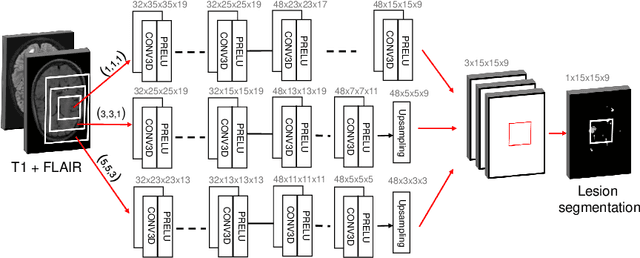
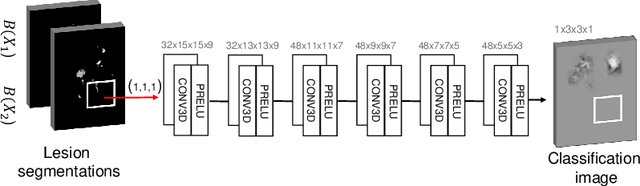
Abstract:The evaluation of white matter lesion progression is an important biomarker in the follow-up of MS patients and plays a crucial role when deciding the course of treatment. Current automated lesion segmentation algorithms are susceptible to variability in image characteristics related to MRI scanner or protocol differences. We propose a model that improves the consistency of MS lesion segmentations in inter-scanner studies. First, we train a CNN base model to approximate the performance of icobrain, an FDA-approved clinically available lesion segmentation software. A discriminator model is then trained to predict if two lesion segmentations are based on scans acquired using the same scanner type or not, achieving a 78% accuracy in this task. Finally, the base model and the discriminator are trained adversarially on multi-scanner longitudinal data to improve the inter-scanner consistency of the base model. The performance of the models is evaluated on an unseen dataset containing manual delineations. The inter-scanner variability is evaluated on test-retest data, where the adversarial network produces improved results over the base model and the FDA-approved solution.
Relevance Vector Machines for harmonization of MRI brain volumes using image descriptors
Nov 08, 2019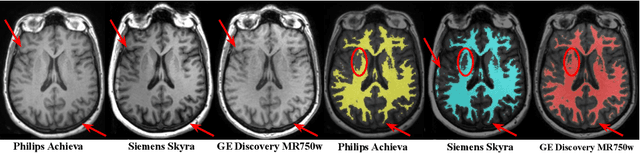
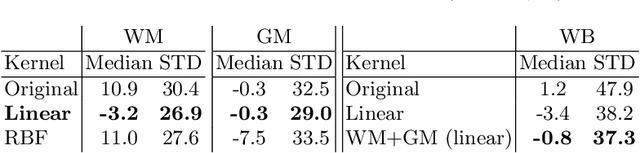
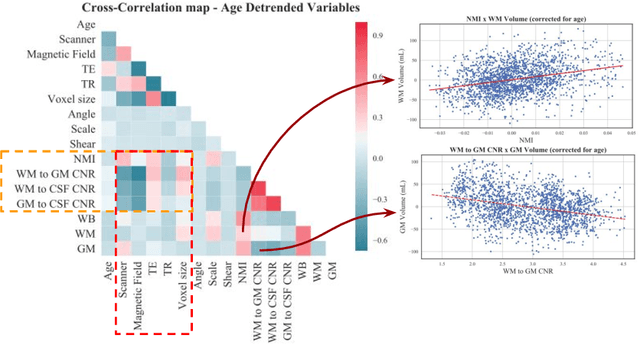
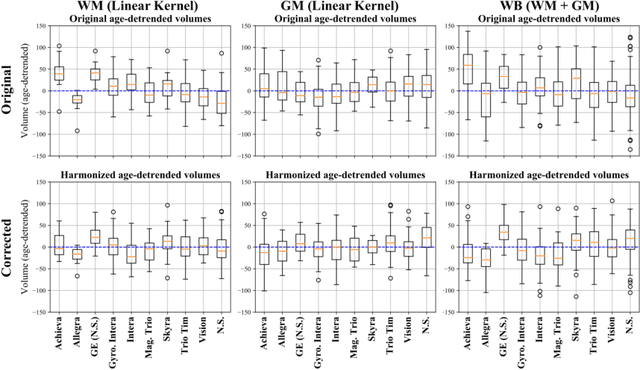
Abstract:With the increased need for multi-center magnetic resonance imaging studies, problems arise related to differences in hardware and software between centers. Namely, current algorithms for brain volume quantification are unreliable for the longitudinal assessment of volume changes in this type of setting. Currently most methods attempt to decrease this issue by regressing the scanner- and/or center-effects from the original data. In this work, we explore a novel approach to harmonize brain volume measurements by using only image descriptors. First, we explore the relationships between volumes and image descriptors. Then, we train a Relevance Vector Machine (RVM) model over a large multi-site dataset of healthy subjects to perform volume harmonization. Finally, we validate the method over two different datasets: i) a subset of unseen healthy controls; and ii) a test-retest dataset of multiple sclerosis (MS) patients. The method decreases scanner and center variability while preserving measurements that did not require correction in MS patient data. We show that image descriptors can be used as input to a machine learning algorithm to improve the reliability of longitudinal volumetric studies.
* 9 pages, 4 figures. Presented at the International Workshop on Machine Learning in Clinical Neuroimaging (MLCN) 2019
A Radiomics Approach to Traumatic Brain Injury Prediction in CT Scans
Nov 14, 2018



Abstract:Computer Tomography (CT) is the gold standard technique for brain damage evaluation after acute Traumatic Brain Injury (TBI). It allows identification of most lesion types and determines the need of surgical or alternative therapeutic procedures. However, the traditional approach for lesion classification is restricted to visual image inspection. In this work, we characterize and predict TBI lesions by using CT-derived radiomics descriptors. Relevant shape, intensity and texture biomarkers characterizing the different lesions are isolated and a lesion predictive model is built by using Partial Least Squares. On a dataset containing 155 scans (105 train, 50 test) the methodology achieved 89.7 % accuracy over the unseen data. When a model was build using only texture features, a 88.2 % accuracy was obtained. Our results suggest that selected radiomics descriptors could play a key role in brain injury prediction. Besides, the proposed methodology is close to reproduce radiologists decision making. These results open new possibilities for radiomics-inspired brain lesion detection, segmentation and prediction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge