Anjany Sekuboyina
Department of Diagnostic and Interventional Neuroradiology, School of Medicine, Klinikum rechts der Isar, Technical University of Munich, Germany, Department of Informatics, Technical University of Munich, Germany
VERIDAH: Solving Enumeration Anomaly Aware Vertebra Labeling across Imaging Sequences
Jan 20, 2026Abstract:The human spine commonly consists of seven cervical, twelve thoracic, and five lumbar vertebrae. However, enumeration anomalies may result in individuals having eleven or thirteen thoracic vertebrae and four or six lumbar vertebrae. Although the identification of enumeration anomalies has potential clinical implications for chronic back pain and operation planning, the thoracolumbar junction is often poorly assessed and rarely described in clinical reports. Additionally, even though multiple deep-learning-based vertebra labeling algorithms exist, there is a lack of methods to automatically label enumeration anomalies. Our work closes that gap by introducing "Vertebra Identification with Anomaly Handling" (VERIDAH), a novel vertebra labeling algorithm based on multiple classification heads combined with a weighted vertebra sequence prediction algorithm. We show that our approach surpasses existing models on T2w TSE sagittal (98.30% vs. 94.24% of subjects with all vertebrae correctly labeled, p < 0.001) and CT imaging (99.18% vs. 77.26% of subjects with all vertebrae correctly labeled, p < 0.001) and works in arbitrary field-of-view images. VERIDAH correctly labeled the presence 2 Möller et al. of thoracic enumeration anomalies in 87.80% and 96.30% of T2w and CT images, respectively, and lumbar enumeration anomalies in 94.48% and 97.22% for T2w and CT, respectively. Our code and models are available at: https://github.com/Hendrik-code/spineps.
DeViDe: Faceted medical knowledge for improved medical vision-language pre-training
Apr 04, 2024Abstract:Vision-language pre-training for chest X-rays has made significant strides, primarily by utilizing paired radiographs and radiology reports. However, existing approaches often face challenges in encoding medical knowledge effectively. While radiology reports provide insights into the current disease manifestation, medical definitions (as used by contemporary methods) tend to be overly abstract, creating a gap in knowledge. To address this, we propose DeViDe, a novel transformer-based method that leverages radiographic descriptions from the open web. These descriptions outline general visual characteristics of diseases in radiographs, and when combined with abstract definitions and radiology reports, provide a holistic snapshot of knowledge. DeViDe incorporates three key features for knowledge-augmented vision language alignment: First, a large-language model-based augmentation is employed to homogenise medical knowledge from diverse sources. Second, this knowledge is aligned with image information at various levels of granularity. Third, a novel projection layer is proposed to handle the complexity of aligning each image with multiple descriptions arising in a multi-label setting. In zero-shot settings, DeViDe performs comparably to fully supervised models on external datasets and achieves state-of-the-art results on three large-scale datasets. Additionally, fine-tuning DeViDe on four downstream tasks and six segmentation tasks showcases its superior performance across data from diverse distributions.
Enhancing Interpretability of Vertebrae Fracture Grading using Human-interpretable Prototypes
Apr 03, 2024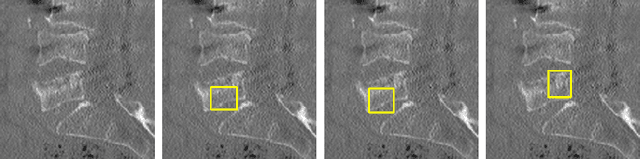
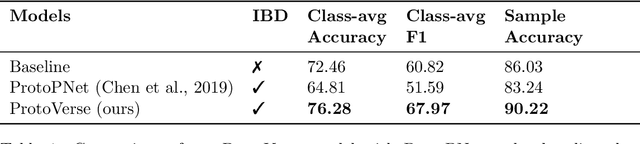
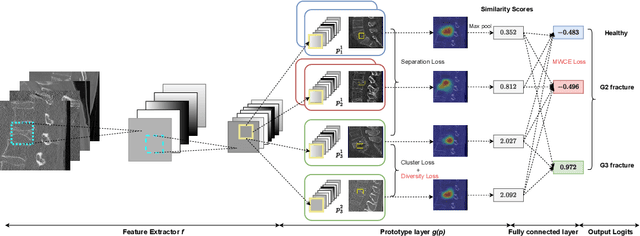

Abstract:Vertebral fracture grading classifies the severity of vertebral fractures, which is a challenging task in medical imaging and has recently attracted Deep Learning (DL) models. Only a few works attempted to make such models human-interpretable despite the need for transparency and trustworthiness in critical use cases like DL-assisted medical diagnosis. Moreover, such models either rely on post-hoc methods or additional annotations. In this work, we propose a novel interpretable-by-design method, ProtoVerse, to find relevant sub-parts of vertebral fractures (prototypes) that reliably explain the model's decision in a human-understandable way. Specifically, we introduce a novel diversity-promoting loss to mitigate prototype repetitions in small datasets with intricate semantics. We have experimented with the VerSe'19 dataset and outperformed the existing prototype-based method. Further, our model provides superior interpretability against the post-hoc method. Importantly, expert radiologists validated the visual interpretability of our results, showing clinical applicability.
A foundation model utilizing chest CT volumes and radiology reports for supervised-level zero-shot detection of abnormalities
Mar 26, 2024Abstract:A major challenge in computational research in 3D medical imaging is the lack of comprehensive datasets. Addressing this issue, our study introduces CT-RATE, the first 3D medical imaging dataset that pairs images with textual reports. CT-RATE consists of 25,692 non-contrast chest CT volumes, expanded to 50,188 through various reconstructions, from 21,304 unique patients, along with corresponding radiology text reports. Leveraging CT-RATE, we developed CT-CLIP, a CT-focused contrastive language-image pre-training framework. As a versatile, self-supervised model, CT-CLIP is designed for broad application and does not require task-specific training. Remarkably, CT-CLIP outperforms state-of-the-art, fully supervised methods in multi-abnormality detection across all key metrics, thus eliminating the need for manual annotation. We also demonstrate its utility in case retrieval, whether using imagery or textual queries, thereby advancing knowledge dissemination. The open-source release of CT-RATE and CT-CLIP marks a significant advancement in medical AI, enhancing 3D imaging analysis and fostering innovation in healthcare.
SPINEPS -- Automatic Whole Spine Segmentation of T2-weighted MR images using a Two-Phase Approach to Multi-class Semantic and Instance Segmentation
Feb 26, 2024
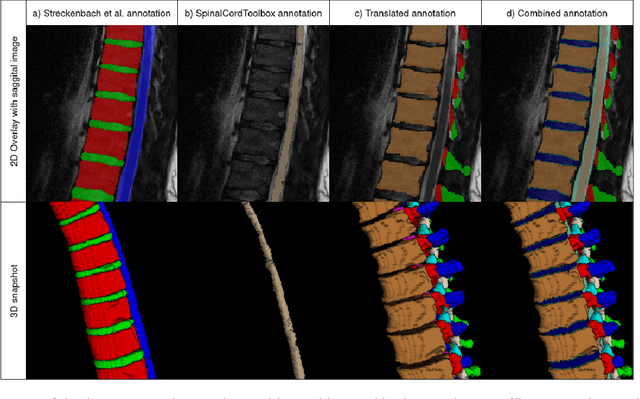
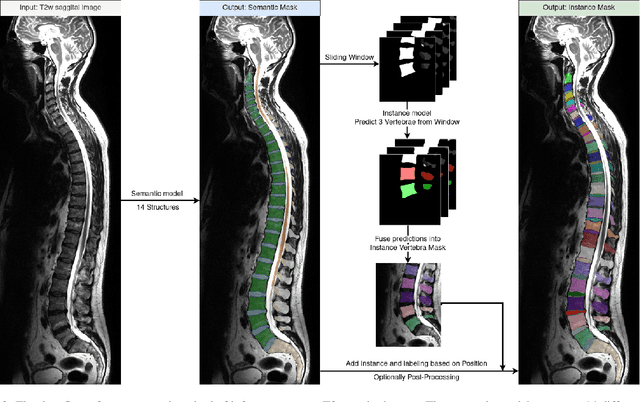
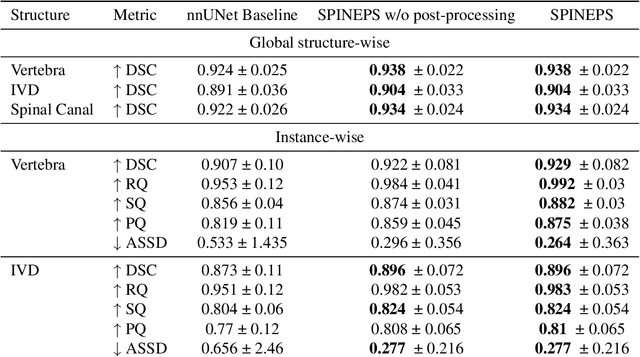
Abstract:Purpose. To present SPINEPS, an open-source deep learning approach for semantic and instance segmentation of 14 spinal structures (ten vertebra substructures, intervertebral discs, spinal cord, spinal canal, and sacrum) in whole body T2w MRI. Methods. During this HIPPA-compliant, retrospective study, we utilized the public SPIDER dataset (218 subjects, 63% female) and a subset of the German National Cohort (1423 subjects, mean age 53, 49% female) for training and evaluation. We combined CT and T2w segmentations to train models that segment 14 spinal structures in T2w sagittal scans both semantically and instance-wise. Performance evaluation metrics included Dice similarity coefficient, average symmetrical surface distance, panoptic quality, segmentation quality, and recognition quality. Statistical significance was assessed using the Wilcoxon signed-rank test. An in-house dataset was used to qualitatively evaluate out-of-distribution samples. Results. On the public dataset, our approach outperformed the baseline (instance-wise vertebra dice score 0.929 vs. 0.907, p-value<0.001). Training on auto-generated annotations and evaluating on manually corrected test data from the GNC yielded global dice scores of 0.900 for vertebrae, 0.960 for intervertebral discs, and 0.947 for the spinal canal. Incorporating the SPIDER dataset during training increased these scores to 0.920, 0.967, 0.958, respectively. Conclusions. The proposed segmentation approach offers robust segmentation of 14 spinal structures in T2w sagittal images, including the spinal cord, spinal canal, intervertebral discs, endplate, sacrum, and vertebrae. The approach yields both a semantic and instance mask as output, thus being easy to utilize. This marks the first publicly available algorithm for whole spine segmentation in sagittal T2w MR imaging.
MultiMedEval: A Benchmark and a Toolkit for Evaluating Medical Vision-Language Models
Feb 16, 2024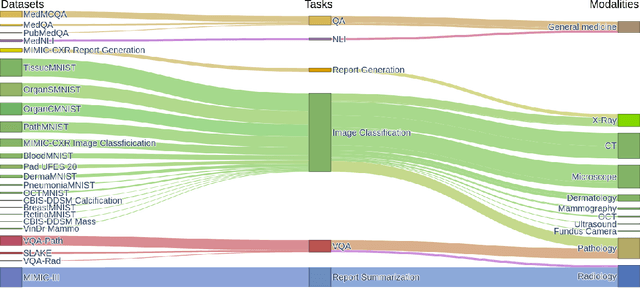
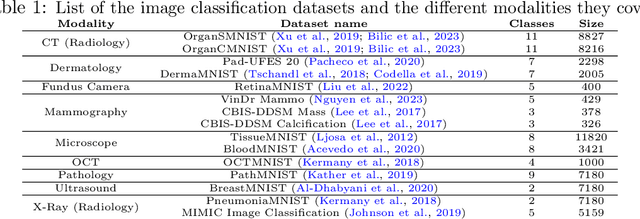
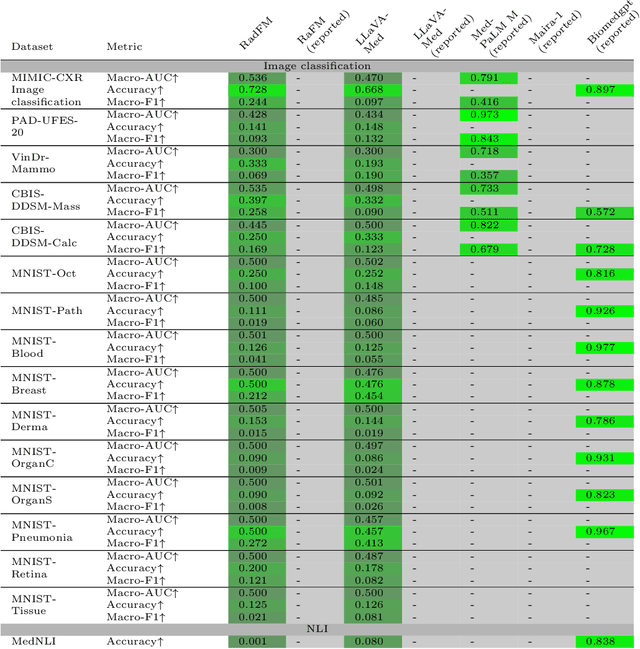
Abstract:We introduce MultiMedEval, an open-source toolkit for fair and reproducible evaluation of large, medical vision-language models (VLM). MultiMedEval comprehensively assesses the models' performance on a broad array of six multi-modal tasks, conducted over 23 datasets, and spanning over 11 medical domains. The chosen tasks and performance metrics are based on their widespread adoption in the community and their diversity, ensuring a thorough evaluation of the model's overall generalizability. We open-source a Python toolkit (github.com/corentin-ryr/MultiMedEval) with a simple interface and setup process, enabling the evaluation of any VLM in just a few lines of code. Our goal is to simplify the intricate landscape of VLM evaluation, thus promoting fair and uniform benchmarking of future models.
Benchmarking the CoW with the TopCoW Challenge: Topology-Aware Anatomical Segmentation of the Circle of Willis for CTA and MRA
Dec 29, 2023



Abstract:The Circle of Willis (CoW) is an important network of arteries connecting major circulations of the brain. Its vascular architecture is believed to affect the risk, severity, and clinical outcome of serious neuro-vascular diseases. However, characterizing the highly variable CoW anatomy is still a manual and time-consuming expert task. The CoW is usually imaged by two angiographic imaging modalities, magnetic resonance angiography (MRA) and computed tomography angiography (CTA), but there exist limited public datasets with annotations on CoW anatomy, especially for CTA. Therefore we organized the TopCoW Challenge in 2023 with the release of an annotated CoW dataset and invited submissions worldwide for the CoW segmentation task, which attracted over 140 registered participants from four continents. TopCoW dataset was the first public dataset with voxel-level annotations for CoW's 13 vessel components, made possible by virtual-reality (VR) technology. It was also the first dataset with paired MRA and CTA from the same patients. TopCoW challenge aimed to tackle the CoW characterization problem as a multiclass anatomical segmentation task with an emphasis on topological metrics. The top performing teams managed to segment many CoW components to Dice scores around 90%, but with lower scores for communicating arteries and rare variants. There were also topological mistakes for predictions with high Dice scores. Additional topological analysis revealed further areas for improvement in detecting certain CoW components and matching CoW variant's topology accurately. TopCoW represented a first attempt at benchmarking the CoW anatomical segmentation task for MRA and CTA, both morphologically and topologically.
MedShapeNet -- A Large-Scale Dataset of 3D Medical Shapes for Computer Vision
Sep 12, 2023



Abstract:We present MedShapeNet, a large collection of anatomical shapes (e.g., bones, organs, vessels) and 3D surgical instrument models. Prior to the deep learning era, the broad application of statistical shape models (SSMs) in medical image analysis is evidence that shapes have been commonly used to describe medical data. Nowadays, however, state-of-the-art (SOTA) deep learning algorithms in medical imaging are predominantly voxel-based. In computer vision, on the contrary, shapes (including, voxel occupancy grids, meshes, point clouds and implicit surface models) are preferred data representations in 3D, as seen from the numerous shape-related publications in premier vision conferences, such as the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), as well as the increasing popularity of ShapeNet (about 51,300 models) and Princeton ModelNet (127,915 models) in computer vision research. MedShapeNet is created as an alternative to these commonly used shape benchmarks to facilitate the translation of data-driven vision algorithms to medical applications, and it extends the opportunities to adapt SOTA vision algorithms to solve critical medical problems. Besides, the majority of the medical shapes in MedShapeNet are modeled directly on the imaging data of real patients, and therefore it complements well existing shape benchmarks comprising of computer-aided design (CAD) models. MedShapeNet currently includes more than 100,000 medical shapes, and provides annotations in the form of paired data. It is therefore also a freely available repository of 3D models for extended reality (virtual reality - VR, augmented reality - AR, mixed reality - MR) and medical 3D printing. This white paper describes in detail the motivations behind MedShapeNet, the shape acquisition procedures, the use cases, as well as the usage of the online shape search portal: https://medshapenet.ikim.nrw/
Denoising diffusion-based MR to CT image translation enables whole spine vertebral segmentation in 2D and 3D without manual annotations
Aug 18, 2023Abstract:Background: Automated segmentation of spinal MR images plays a vital role both scientifically and clinically. However, accurately delineating posterior spine structures presents challenges. Methods: This retrospective study, approved by the ethical committee, involved translating T1w and T2w MR image series into CT images in a total of n=263 pairs of CT/MR series. Landmark-based registration was performed to align image pairs. We compared 2D paired (Pix2Pix, denoising diffusion implicit models (DDIM) image mode, DDIM noise mode) and unpaired (contrastive unpaired translation, SynDiff) image-to-image translation using "peak signal to noise ratio" (PSNR) as quality measure. A publicly available segmentation network segmented the synthesized CT datasets, and Dice scores were evaluated on in-house test sets and the "MRSpineSeg Challenge" volumes. The 2D findings were extended to 3D Pix2Pix and DDIM. Results: 2D paired methods and SynDiff exhibited similar translation performance and Dice scores on paired data. DDIM image mode achieved the highest image quality. SynDiff, Pix2Pix, and DDIM image mode demonstrated similar Dice scores (0.77). For craniocaudal axis rotations, at least two landmarks per vertebra were required for registration. The 3D translation outperformed the 2D approach, resulting in improved Dice scores (0.80) and anatomically accurate segmentations in a higher resolution than the original MR image. Conclusion: Two landmarks per vertebra registration enabled paired image-to-image translation from MR to CT and outperformed all unpaired approaches. The 3D techniques provided anatomically correct segmentations, avoiding underprediction of small structures like the spinous process.
Diffusion-Based Hierarchical Multi-Label Object Detection to Analyze Panoramic Dental X-rays
Mar 20, 2023



Abstract:Due to the necessity for precise treatment planning, the use of panoramic X-rays to identify different dental diseases has tremendously increased. Although numerous ML models have been developed for the interpretation of panoramic X-rays, there has not been an end-to-end model developed that can identify problematic teeth with dental enumeration and associated diagnoses at the same time. To develop such a model, we structure the three distinct types of annotated data hierarchically following the FDI system, the first labeled with only quadrant, the second labeled with quadrant-enumeration, and the third fully labeled with quadrant-enumeration-diagnosis. To learn from all three hierarchies jointly, we introduce a novel diffusion-based hierarchical multi-label object detection framework by adapting a diffusion-based method that formulates object detection as a denoising diffusion process from noisy boxes to object boxes. Specifically, to take advantage of the hierarchically annotated data, our method utilizes a novel noisy box manipulation technique by adapting the denoising process in the diffusion network with the inference from the previously trained model in hierarchical order. We also utilize a multi-label object detection method to learn efficiently from partial annotations and to give all the needed information about each abnormal tooth for treatment planning. Experimental results show that our method significantly outperforms state-of-the-art object detection methods, including RetinaNet, Faster R-CNN, DETR, and DiffusionDet for the analysis of panoramic X-rays, demonstrating the great potential of our method for hierarchically and partially annotated datasets. The code and the data are available at: https://github.com/ibrahimethemhamamci/HierarchicalDet.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge