Bastian Wittmann
vesselFM: A Foundation Model for Universal 3D Blood Vessel Segmentation
Nov 26, 2024



Abstract:Segmenting 3D blood vessels is a critical yet challenging task in medical image analysis. This is due to significant imaging modality-specific variations in artifacts, vascular patterns and scales, signal-to-noise ratios, and background tissues. These variations, along with domain gaps arising from varying imaging protocols, limit the generalization of existing supervised learning-based methods, requiring tedious voxel-level annotations for each dataset separately. While foundation models promise to alleviate this limitation, they typically fail to generalize to the task of blood vessel segmentation, posing a unique, complex problem. In this work, we present vesselFM, a foundation model designed specifically for the broad task of 3D blood vessel segmentation. Unlike previous models, vesselFM can effortlessly generalize to unseen domains. To achieve zero-shot generalization, we train vesselFM on three heterogeneous data sources: a large, curated annotated dataset, data generated by a domain randomization scheme, and data sampled from a flow matching-based generative model. Extensive evaluations show that vesselFM outperforms state-of-the-art medical image segmentation foundation models across four (pre-)clinically relevant imaging modalities in zero-, one-, and few-shot scenarios, therefore providing a universal solution for 3D blood vessel segmentation.
Predicting Stroke through Retinal Graphs and Multimodal Self-supervised Learning
Nov 08, 2024



Abstract:Early identification of stroke is crucial for intervention, requiring reliable models. We proposed an efficient retinal image representation together with clinical information to capture a comprehensive overview of cardiovascular health, leveraging large multimodal datasets for new medical insights. Our approach is one of the first contrastive frameworks that integrates graph and tabular data, using vessel graphs derived from retinal images for efficient representation. This method, combined with multimodal contrastive learning, significantly enhances stroke prediction accuracy by integrating data from multiple sources and using contrastive learning for transfer learning. The self-supervised learning techniques employed allow the model to learn effectively from unlabeled data, reducing the dependency on large annotated datasets. Our framework showed an AUROC improvement of 3.78% from supervised to self-supervised approaches. Additionally, the graph-level representation approach achieved superior performance to image encoders while significantly reducing pre-training and fine-tuning runtimes. These findings indicate that retinal images are a cost-effective method for improving cardiovascular disease predictions and pave the way for future research into retinal and cerebral vessel connections and the use of graph-based retinal vessel representations.
A foundation model utilizing chest CT volumes and radiology reports for supervised-level zero-shot detection of abnormalities
Mar 26, 2024Abstract:A major challenge in computational research in 3D medical imaging is the lack of comprehensive datasets. Addressing this issue, our study introduces CT-RATE, the first 3D medical imaging dataset that pairs images with textual reports. CT-RATE consists of 25,692 non-contrast chest CT volumes, expanded to 50,188 through various reconstructions, from 21,304 unique patients, along with corresponding radiology text reports. Leveraging CT-RATE, we developed CT-CLIP, a CT-focused contrastive language-image pre-training framework. As a versatile, self-supervised model, CT-CLIP is designed for broad application and does not require task-specific training. Remarkably, CT-CLIP outperforms state-of-the-art, fully supervised methods in multi-abnormality detection across all key metrics, thus eliminating the need for manual annotation. We also demonstrate its utility in case retrieval, whether using imagery or textual queries, thereby advancing knowledge dissemination. The open-source release of CT-RATE and CT-CLIP marks a significant advancement in medical AI, enhancing 3D imaging analysis and fostering innovation in healthcare.
Simulation-Based Segmentation of Blood Vessels in Cerebral 3D OCTA Images
Mar 11, 2024



Abstract:Segmentation of blood vessels in murine cerebral 3D OCTA images is foundational for in vivo quantitative analysis of the effects of neurovascular disorders, such as stroke or Alzheimer's, on the vascular network. However, to accurately segment blood vessels with state-of-the-art deep learning methods, a vast amount of voxel-level annotations is required. Since cerebral 3D OCTA images are typically plagued by artifacts and generally have a low signal-to-noise ratio, acquiring manual annotations poses an especially cumbersome and time-consuming task. To alleviate the need for manual annotations, we propose utilizing synthetic data to supervise segmentation algorithms. To this end, we extract patches from vessel graphs and transform them into synthetic cerebral 3D OCTA images paired with their matching ground truth labels by simulating the most dominant 3D OCTA artifacts. In extensive experiments, we demonstrate that our approach achieves competitive results, enabling annotation-free blood vessel segmentation in cerebral 3D OCTA images.
Link Prediction for Flow-Driven Spatial Networks
Mar 25, 2023Abstract:Link prediction algorithms predict the existence of connections between nodes in network-structured data and are typically applied to refine the connectivity among nodes by proposing meaningful new links. In this work, we focus on link prediction for flow-driven spatial networks, which are embedded in a Euclidean space and relate to physical exchange and transportation processes (e.g., blood flow in vessels or traffic flow in road networks). To this end, we propose the Graph Attentive Vectors (GAV) link prediction framework. GAV models simplified dynamics of physical flow in spatial networks via an attentive, neighborhood-aware message-passing paradigm, updating vector embeddings in a constrained manner. We evaluate GAV on eight flow-driven spatial networks given by whole-brain vessel graphs and road networks. GAV demonstrates superior performances across all datasets and metrics and outperforms the current state-of-the-art on the ogbl-vessel benchmark by more than 18% (98.38 vs. 83.07 AUC).
Focused Decoding Enables 3D Anatomical Detection by Transformers
Jul 21, 2022

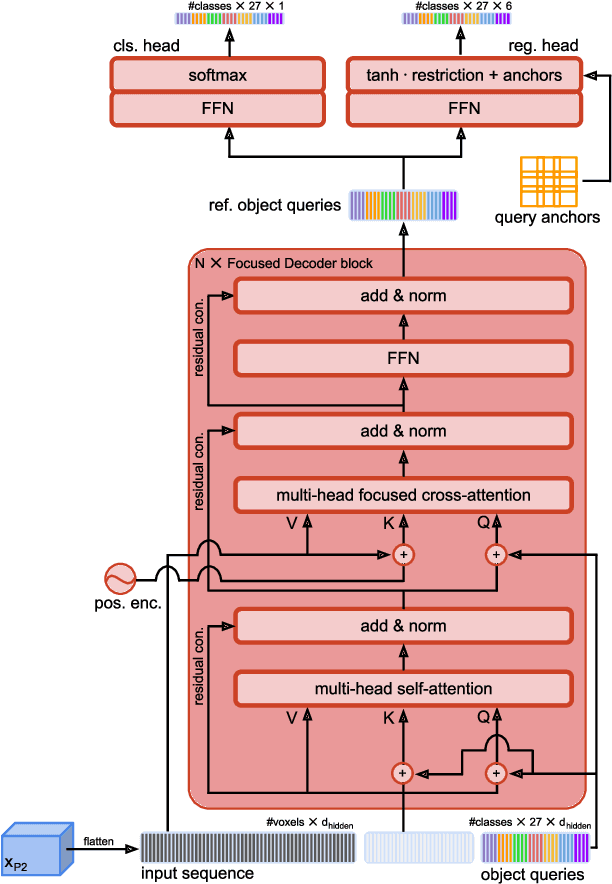

Abstract:Detection Transformers represent end-to-end object detection approaches based on a Transformer encoder-decoder architecture, exploiting the attention mechanism for global relation modeling. Although Detection Transformers deliver results on par with or even superior to their highly optimized CNN-based counterparts operating on 2D natural images, their success is closely coupled to access to a vast amount of training data. This, however, restricts the feasibility of employing Detection Transformers in the medical domain, as access to annotated data is typically limited. To tackle this issue and facilitate the advent of medical Detection Transformers, we propose a novel Detection Transformer for 3D anatomical structure detection, dubbed Focused Decoder. Focused Decoder leverages information from an anatomical region atlas to simultaneously deploy query anchors and restrict the cross-attention's field of view to regions of interest, which allows for a precise focus on relevant anatomical structures. We evaluate our proposed approach on two publicly available CT datasets and demonstrate that Focused Decoder not only provides strong detection results and thus alleviates the need for a vast amount of annotated data but also exhibits exceptional and highly intuitive explainability of results via attention weights. Code for Focused Decoder is available in our medical Vision Transformer library github.com/bwittmann/transoar.
Relationformer: A Unified Framework for Image-to-Graph Generation
Mar 19, 2022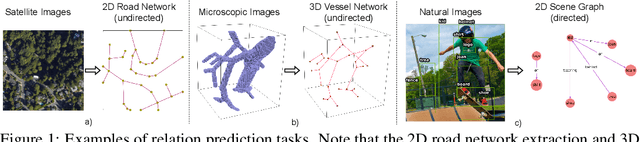

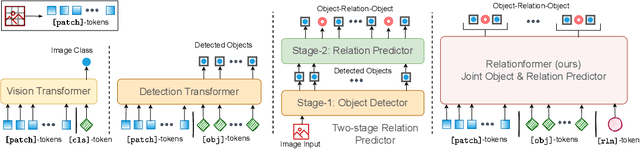
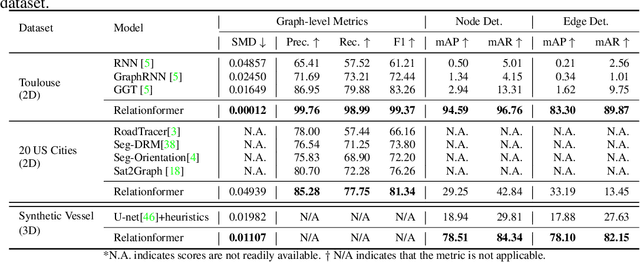
Abstract:A comprehensive representation of an image requires understanding objects and their mutual relationship, especially in image-to-graph generation, e.g., road network extraction, blood-vessel network extraction, or scene graph generation. Traditionally, image-to-graph generation is addressed with a two-stage approach consisting of object detection followed by a separate relation prediction, which prevents simultaneous object-relation interaction. This work proposes a unified one-stage transformer-based framework, namely Relationformer, that jointly predicts objects and their relations. We leverage direct set-based object prediction and incorporate the interaction among the objects to learn an object-relation representation jointly. In addition to existing [obj]-tokens, we propose a novel learnable token, namely [rln]-token. Together with [obj]-tokens, [rln]-token exploits local and global semantic reasoning in an image through a series of mutual associations. In combination with the pair-wise [obj]-token, the [rln]-token contributes to a computationally efficient relation prediction. We achieve state-of-the-art performance on multiple, diverse and multi-domain datasets that demonstrate our approach's effectiveness and generalizability.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge