Daniel Razansky
Simulation-Based Segmentation of Blood Vessels in Cerebral 3D OCTA Images
Mar 11, 2024



Abstract:Segmentation of blood vessels in murine cerebral 3D OCTA images is foundational for in vivo quantitative analysis of the effects of neurovascular disorders, such as stroke or Alzheimer's, on the vascular network. However, to accurately segment blood vessels with state-of-the-art deep learning methods, a vast amount of voxel-level annotations is required. Since cerebral 3D OCTA images are typically plagued by artifacts and generally have a low signal-to-noise ratio, acquiring manual annotations poses an especially cumbersome and time-consuming task. To alleviate the need for manual annotations, we propose utilizing synthetic data to supervise segmentation algorithms. To this end, we extract patches from vessel graphs and transform them into synthetic cerebral 3D OCTA images paired with their matching ground truth labels by simulating the most dominant 3D OCTA artifacts. In extensive experiments, we demonstrate that our approach achieves competitive results, enabling annotation-free blood vessel segmentation in cerebral 3D OCTA images.
OADAT: Experimental and Synthetic Clinical Optoacoustic Data for Standardized Image Processing
Jun 17, 2022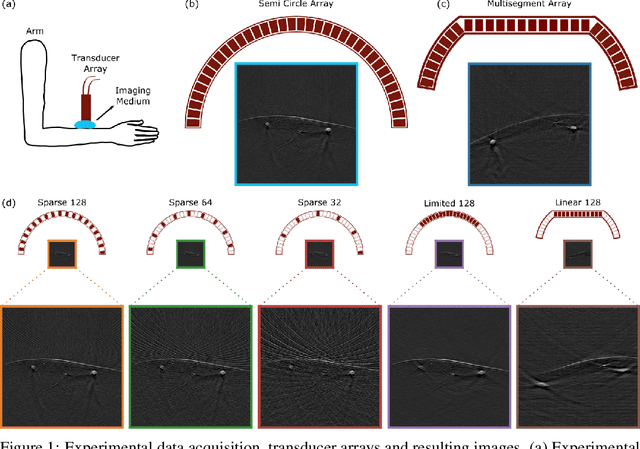
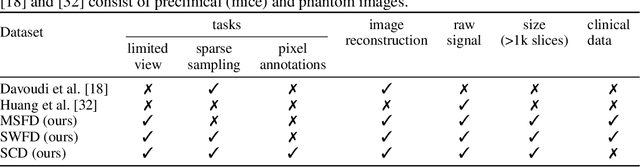
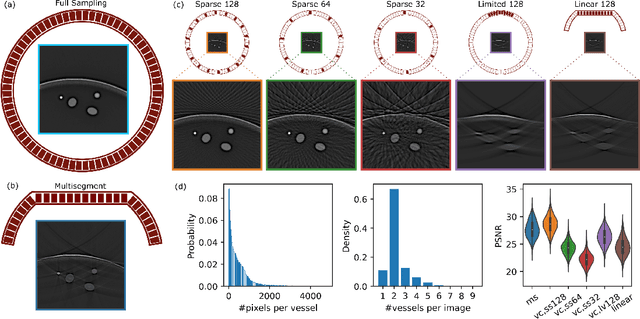
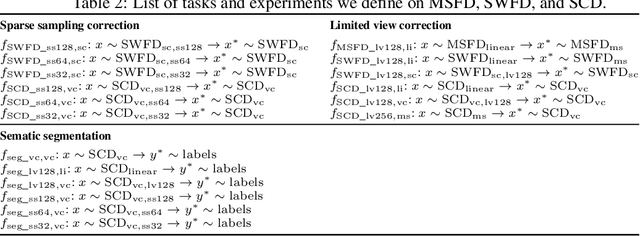
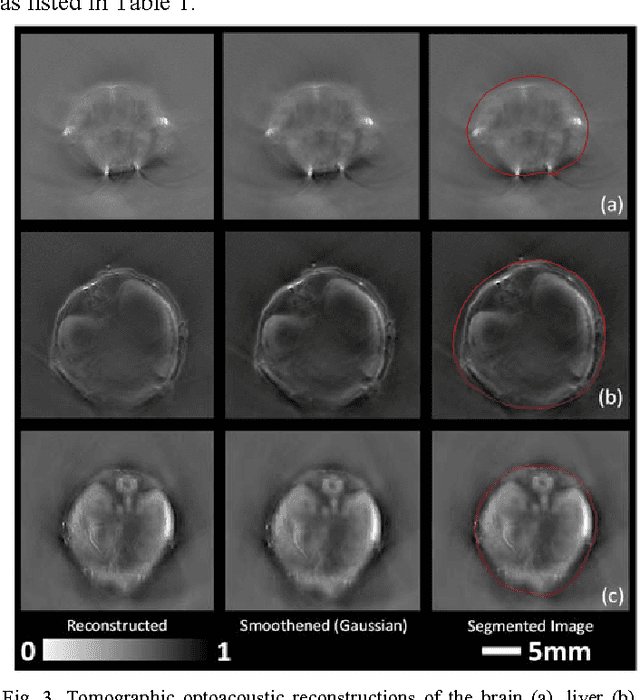
Abstract:Optoacoustic (OA) imaging is based on excitation of biological tissues with nanosecond-duration laser pulses followed by subsequent detection of ultrasound waves generated via light-absorption-mediated thermoelastic expansion. OA imaging features a powerful combination between rich optical contrast and high resolution in deep tissues. This enabled the exploration of a number of attractive new applications both in clinical and laboratory settings. However, no standardized datasets generated with different types of experimental set-up and associated processing methods are available to facilitate advances in broader applications of OA in clinical settings. This complicates an objective comparison between new and established data processing methods, often leading to qualitative results and arbitrary interpretations of the data. In this paper, we provide both experimental and synthetic OA raw signals and reconstructed image domain datasets rendered with different experimental parameters and tomographic acquisition geometries. We further provide trained neural networks to tackle three important challenges related to OA image processing, namely accurate reconstruction under limited view tomographic conditions, removal of spatial undersampling artifacts and anatomical segmentation for improved image reconstruction. Specifically, we define 18 experiments corresponding to the aforementioned challenges as benchmarks to be used as a reference for the development of more advanced processing methods.
Deep learning facilitates fully automated brain image registration of optoacoustic tomography and magnetic resonance imaging
Sep 04, 2021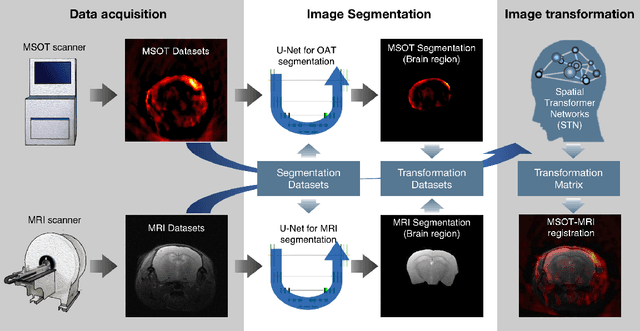
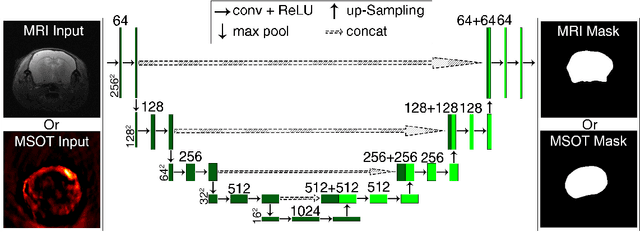
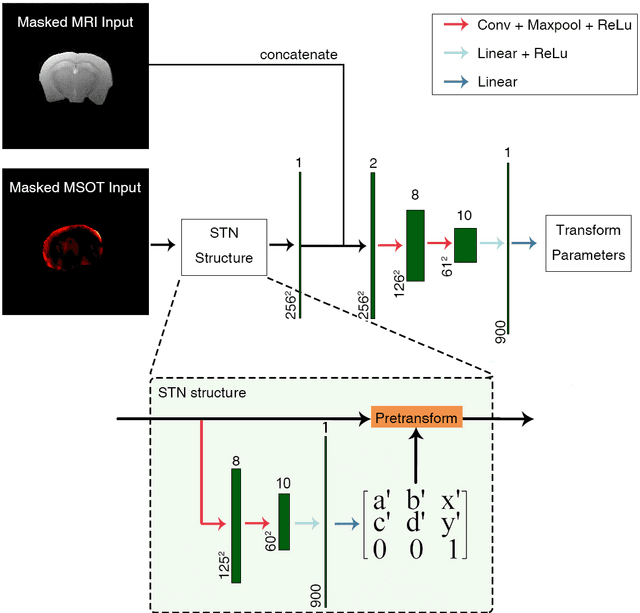

Abstract:Multi-spectral optoacoustic tomography (MSOT) is an emerging optical imaging method providing multiplex molecular and functional information from the rodent brain. It can be greatly augmented by magnetic resonance imaging (MRI) that offers excellent soft-tissue contrast and high-resolution brain anatomy. Nevertheless, registration of multi-modal images remains challenging, chiefly due to the entirely different image contrast rendered by these modalities. Previously reported registration algorithms mostly relied on manual user-dependent brain segmentation, which compromised data interpretation and accurate quantification. Here we propose a fully automated registration method for MSOT-MRI multimodal imaging empowered by deep learning. The automated workflow includes neural network-based image segmentation to generate suitable masks, which are subsequently registered using an additional neural network. Performance of the algorithm is showcased with datasets acquired by cross-sectional MSOT and high-field MRI preclinical scanners. The automated registration method is further validated with manual and half-automated registration, demonstrating its robustness and accuracy.
Maximum entropy based non-negative optoacoustic tomographic image reconstruction
Jul 26, 2017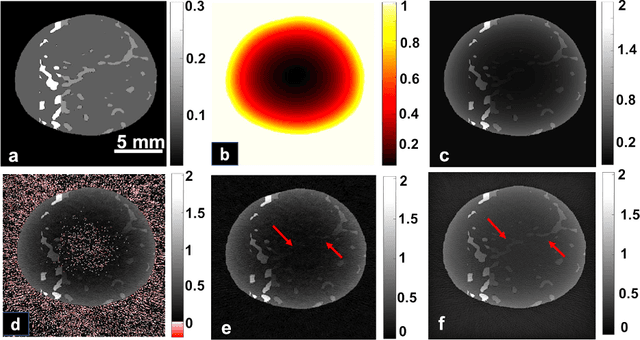
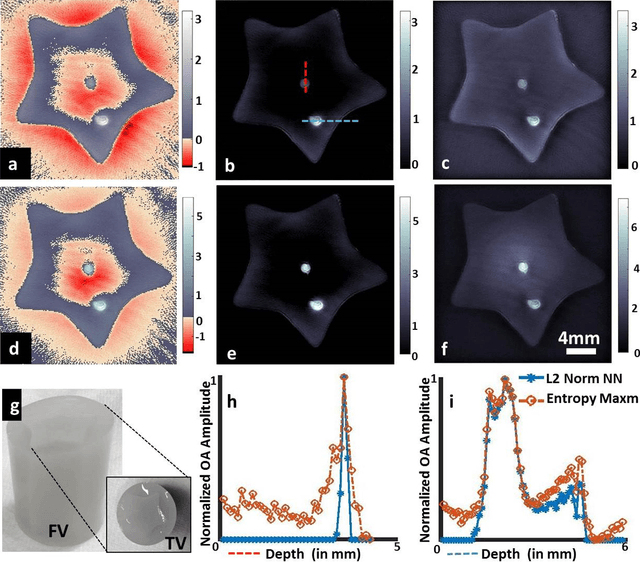
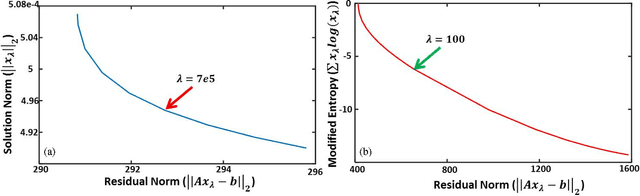
Abstract:Optoacoustic (photoacoustic) tomography reconstructs maps of the initial pressure rise induced by the absorption of light pulses in tissue. In practice, due to inaccurate assumptions in the forward model employed, noise and other experimental factors, the images often contain errors, occasionally manifested as negative values. We present optoacoustic tomography based on an entropy maximization algorithm that uses logarithmic regularization as a potent method for imparting non-negative image reconstruction. We experimentally investigate the performance achieved by the entropy maximization scheme on phantoms and in vivo samples. The findings demonstrate that the proposed scheme reconstructs physically relevant image values devoid of unwanted negative contrast, thus improving quantitative imaging performance.
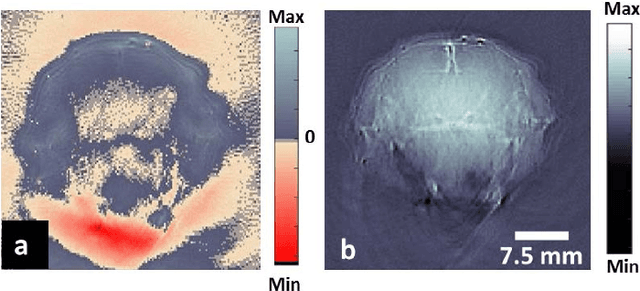
Visual Quality Enhancement in Optoacoustic Tomography using Active Contour Segmentation Priors
Apr 11, 2016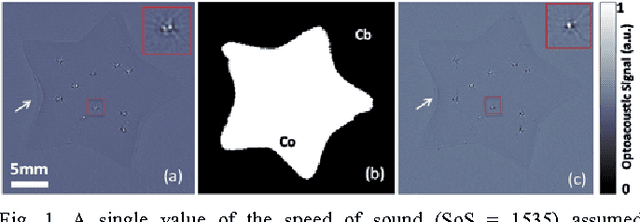
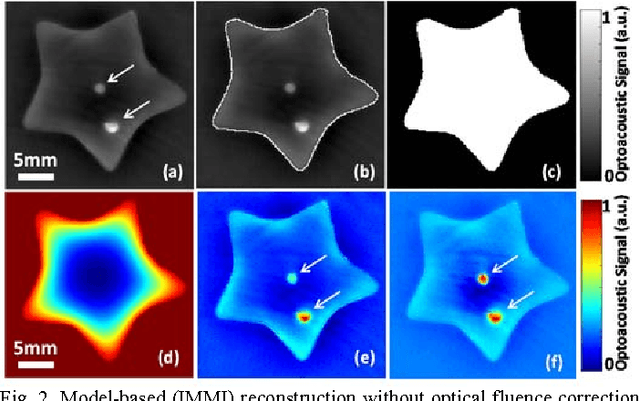
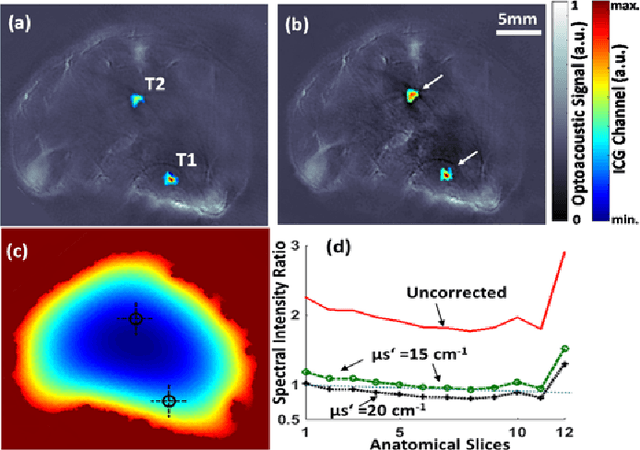
Abstract:Segmentation of biomedical images is essential for studying and characterizing anatomical structures, detection and evaluation of pathological tissues. Segmentation has been further shown to enhance the reconstruction performance in many tomographic imaging modalities by accounting for heterogeneities of the excitation field and tissue properties in the imaged region. This is particularly relevant in optoacoustic tomography, where discontinuities in the optical and acoustic tissue properties, if not properly accounted for, may result in deterioration of the imaging performance. Efficient segmentation of optoacoustic images is often hampered by the relatively low intrinsic contrast of large anatomical structures, which is further impaired by the limited angular coverage of some commonly employed tomographic imaging configurations. Herein, we analyze the performance of active contour models for boundary segmentation in cross-sectional optoacoustic tomography. The segmented mask is employed to construct a two compartment model for the acoustic and optical parameters of the imaged tissues, which is subsequently used to improve accuracy of the image reconstruction routines. The performance of the suggested segmentation and modeling approach are showcased in tissue-mimicking phantoms and small animal imaging experiments.
Multiscale edge detection and parametric shape modeling for boundary delineation in optoacoustic images
Jun 09, 2015

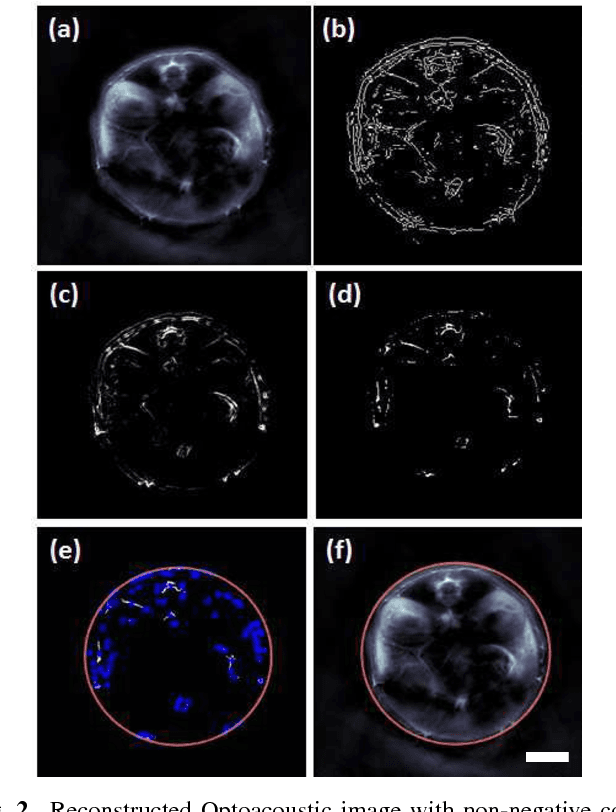
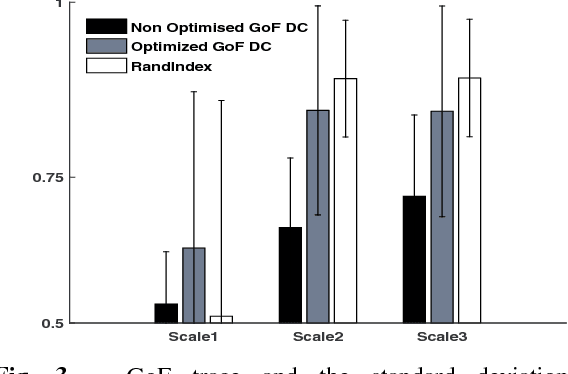
Abstract:In this article, we present a novel scheme for segmenting the image boundary (with the background) in optoacoustic small animal in vivo imaging systems. The method utilizes a multiscale edge detection algorithm to generate a binary edge map. A scale dependent morphological operation is employed to clean spurious edges. Thereafter, an ellipse is fitted to the edge map through constrained parametric transformations and iterative goodness of fit calculations. The method delimits the tissue edges through the curve fitting model, which has shown high levels of accuracy. Thus, this method enables segmentation of optoacoutic images with minimal human intervention, by eliminating need of scale selection for multiscale processing and seed point determination for contour mapping.
* Engineering in Medicine and Biology Society (EMBC), 2015 37th Annual International Conference of the IEEE (Accepted version)
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge