Berkan Lafci
A foundation model utilizing chest CT volumes and radiology reports for supervised-level zero-shot detection of abnormalities
Mar 26, 2024Abstract:A major challenge in computational research in 3D medical imaging is the lack of comprehensive datasets. Addressing this issue, our study introduces CT-RATE, the first 3D medical imaging dataset that pairs images with textual reports. CT-RATE consists of 25,692 non-contrast chest CT volumes, expanded to 50,188 through various reconstructions, from 21,304 unique patients, along with corresponding radiology text reports. Leveraging CT-RATE, we developed CT-CLIP, a CT-focused contrastive language-image pre-training framework. As a versatile, self-supervised model, CT-CLIP is designed for broad application and does not require task-specific training. Remarkably, CT-CLIP outperforms state-of-the-art, fully supervised methods in multi-abnormality detection across all key metrics, thus eliminating the need for manual annotation. We also demonstrate its utility in case retrieval, whether using imagery or textual queries, thereby advancing knowledge dissemination. The open-source release of CT-RATE and CT-CLIP marks a significant advancement in medical AI, enhancing 3D imaging analysis and fostering innovation in healthcare.
OADAT: Experimental and Synthetic Clinical Optoacoustic Data for Standardized Image Processing
Jun 17, 2022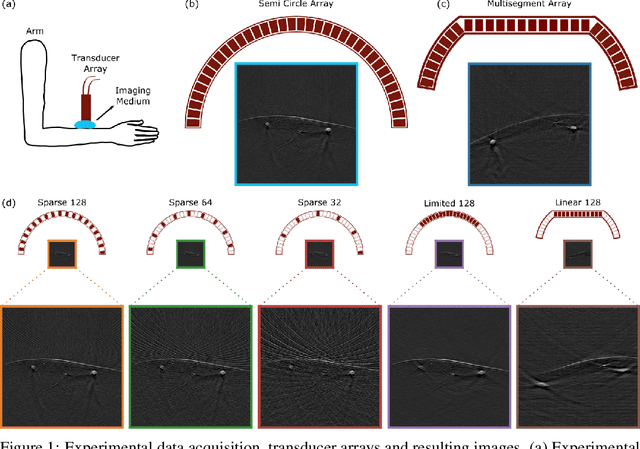
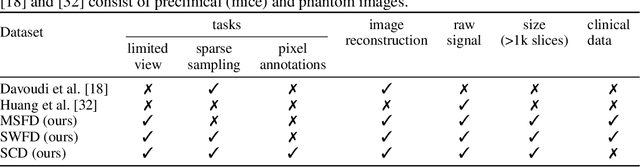
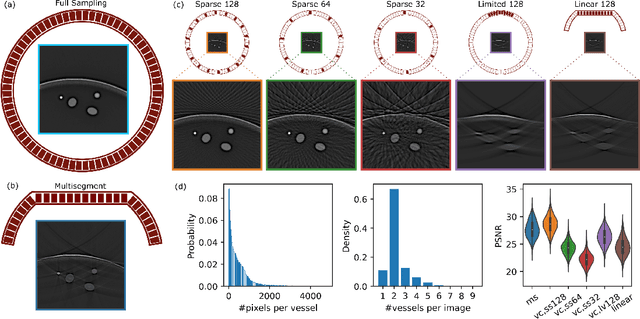
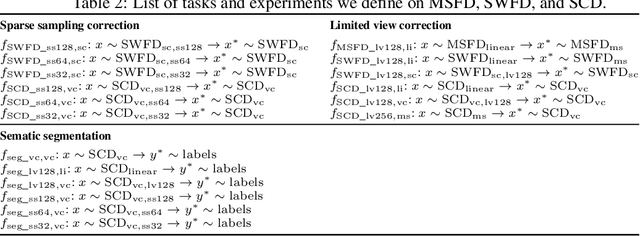
Abstract:Optoacoustic (OA) imaging is based on excitation of biological tissues with nanosecond-duration laser pulses followed by subsequent detection of ultrasound waves generated via light-absorption-mediated thermoelastic expansion. OA imaging features a powerful combination between rich optical contrast and high resolution in deep tissues. This enabled the exploration of a number of attractive new applications both in clinical and laboratory settings. However, no standardized datasets generated with different types of experimental set-up and associated processing methods are available to facilitate advances in broader applications of OA in clinical settings. This complicates an objective comparison between new and established data processing methods, often leading to qualitative results and arbitrary interpretations of the data. In this paper, we provide both experimental and synthetic OA raw signals and reconstructed image domain datasets rendered with different experimental parameters and tomographic acquisition geometries. We further provide trained neural networks to tackle three important challenges related to OA image processing, namely accurate reconstruction under limited view tomographic conditions, removal of spatial undersampling artifacts and anatomical segmentation for improved image reconstruction. Specifically, we define 18 experiments corresponding to the aforementioned challenges as benchmarks to be used as a reference for the development of more advanced processing methods.
Deep learning facilitates fully automated brain image registration of optoacoustic tomography and magnetic resonance imaging
Sep 04, 2021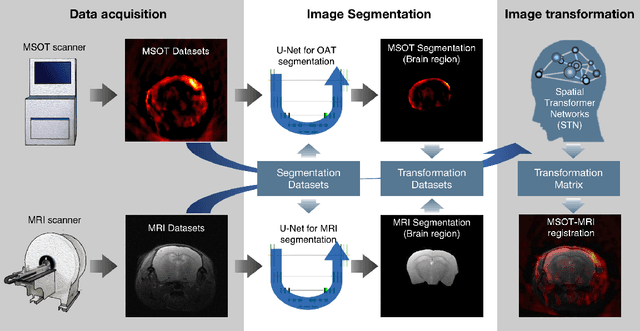
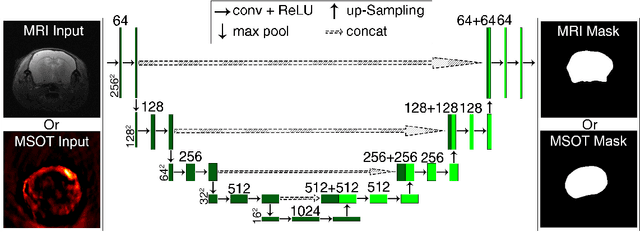
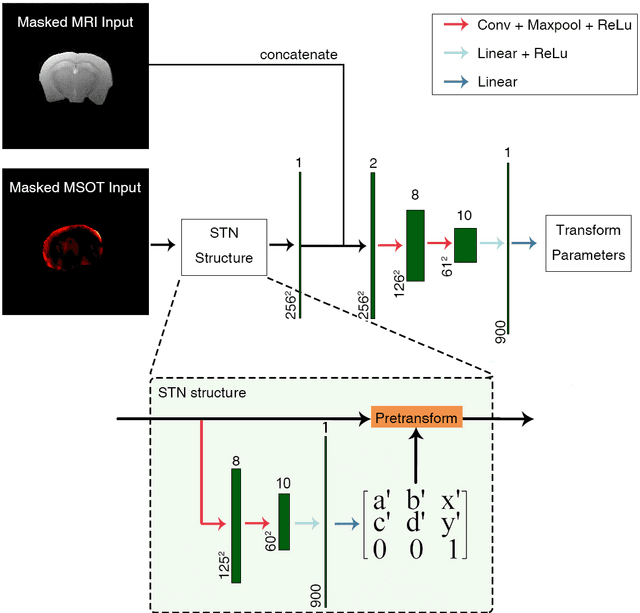

Abstract:Multi-spectral optoacoustic tomography (MSOT) is an emerging optical imaging method providing multiplex molecular and functional information from the rodent brain. It can be greatly augmented by magnetic resonance imaging (MRI) that offers excellent soft-tissue contrast and high-resolution brain anatomy. Nevertheless, registration of multi-modal images remains challenging, chiefly due to the entirely different image contrast rendered by these modalities. Previously reported registration algorithms mostly relied on manual user-dependent brain segmentation, which compromised data interpretation and accurate quantification. Here we propose a fully automated registration method for MSOT-MRI multimodal imaging empowered by deep learning. The automated workflow includes neural network-based image segmentation to generate suitable masks, which are subsequently registered using an additional neural network. Performance of the algorithm is showcased with datasets acquired by cross-sectional MSOT and high-field MRI preclinical scanners. The automated registration method is further validated with manual and half-automated registration, demonstrating its robustness and accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge