Firat Ozdemir
Finetuning a Weather Foundation Model with Lightweight Decoders for Unseen Physical Processes
Jun 23, 2025Abstract:Recent advances in AI weather forecasting have led to the emergence of so-called "foundation models", typically defined by expensive pretraining and minimal fine-tuning for downstream tasks. However, in the natural sciences, a desirable foundation model should also encode meaningful statistical relationships between the underlying physical variables. This study evaluates the performance of the state-of-the-art Aurora foundation model in predicting hydrological variables, which were not considered during pretraining. We introduce a lightweight approach using shallow decoders trained on the latent representations of the pretrained model to predict these new variables. As a baseline, we compare this to fine-tuning the full model, which allows further optimization of the latent space while incorporating new variables into both inputs and outputs. The decoder-based approach requires 50% less training time and 35% less memory, while achieving strong accuracy across various hydrological variables and preserving desirable properties of the foundation model, such as autoregressive stability. Notably, decoder accuracy depends on the physical correlation between the new variables and those used during pretraining, indicating that Aurora's latent space captures meaningful physical relationships. In this sense, we argue that an important quality metric for foundation models in Earth sciences is their ability to be extended to new variables without a full fine-tuning. This provides a new perspective for making foundation models more accessible to communities with limited computational resources, while supporting broader adoption in Earth sciences.
FocusDD: Real-World Scene Infusion for Robust Dataset Distillation
Jan 11, 2025



Abstract:Dataset distillation has emerged as a strategy to compress real-world datasets for efficient training. However, it struggles with large-scale and high-resolution datasets, limiting its practicality. This paper introduces a novel resolution-independent dataset distillation method Focus ed Dataset Distillation (FocusDD), which achieves diversity and realism in distilled data by identifying key information patches, thereby ensuring the generalization capability of the distilled dataset across different network architectures. Specifically, FocusDD leverages a pre-trained Vision Transformer (ViT) to extract key image patches, which are then synthesized into a single distilled image. These distilled images, which capture multiple targets, are suitable not only for classification tasks but also for dense tasks such as object detection. To further improve the generalization of the distilled dataset, each synthesized image is augmented with a downsampled view of the original image. Experimental results on the ImageNet-1K dataset demonstrate that, with 100 images per class (IPC), ResNet50 and MobileNet-v2 achieve validation accuracies of 71.0% and 62.6%, respectively, outperforming state-of-the-art methods by 2.8% and 4.7%. Notably, FocusDD is the first method to use distilled datasets for object detection tasks. On the COCO2017 dataset, with an IPC of 50, YOLOv11n and YOLOv11s achieve 24.4% and 32.1% mAP, respectively, further validating the effectiveness of our approach.
PokeFlex: A Real-World Dataset of Deformable Objects for Robotics
Oct 10, 2024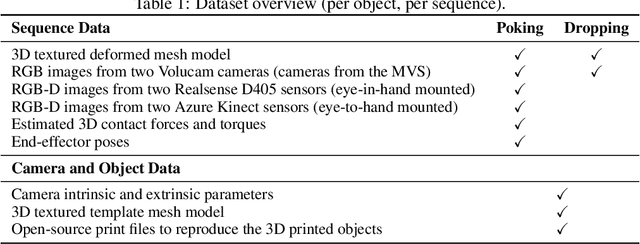
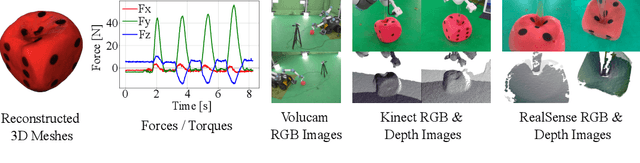
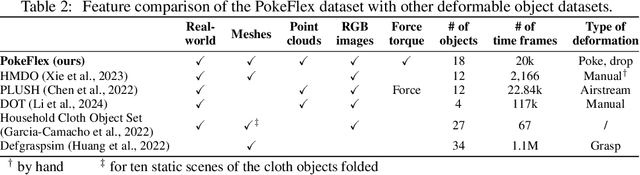

Abstract:Data-driven methods have shown great potential in solving challenging manipulation tasks, however, their application in the domain of deformable objects has been constrained, in part, by the lack of data. To address this, we propose PokeFlex, a dataset featuring real-world paired and annotated multimodal data that includes 3D textured meshes, point clouds, RGB images, and depth maps. Such data can be leveraged for several downstream tasks such as online 3D mesh reconstruction, and it can potentially enable underexplored applications such as the real-world deployment of traditional control methods based on mesh simulations. To deal with the challenges posed by real-world 3D mesh reconstruction, we leverage a professional volumetric capture system that allows complete 360{\deg} reconstruction. PokeFlex consists of 18 deformable objects with varying stiffness and shapes. Deformations are generated by dropping objects onto a flat surface or by poking the objects with a robot arm. Interaction forces and torques are also reported for the latter case. Using different data modalities, we demonstrated a use case for the PokeFlex dataset in online 3D mesh reconstruction. We refer the reader to our website ( https://pokeflex-dataset.github.io/ ) for demos and examples of our dataset.
Retrospective Uncertainties for Deep Models using Vine Copulas
Feb 24, 2023Abstract:Despite the major progress of deep models as learning machines, uncertainty estimation remains a major challenge. Existing solutions rely on modified loss functions or architectural changes. We propose to compensate for the lack of built-in uncertainty estimates by supplementing any network, retrospectively, with a subsequent vine copula model, in an overall compound we call Vine-Copula Neural Network (VCNN). Through synthetic and real-data experiments, we show that VCNNs could be task (regression/classification) and architecture (recurrent, fully connected) agnostic while providing reliable and better-calibrated uncertainty estimates, comparable to state-of-the-art built-in uncertainty solutions.
OADAT: Experimental and Synthetic Clinical Optoacoustic Data for Standardized Image Processing
Jun 17, 2022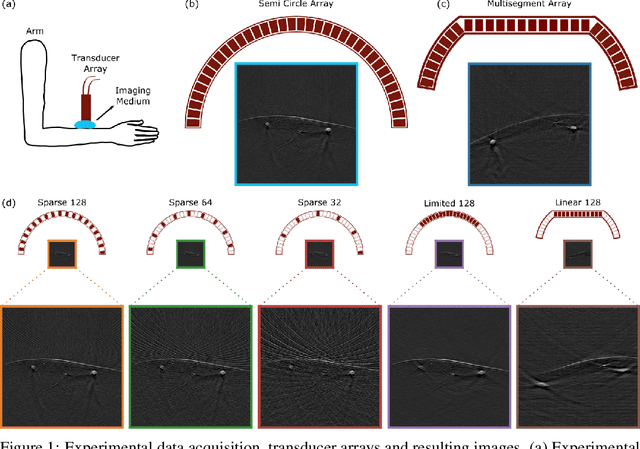
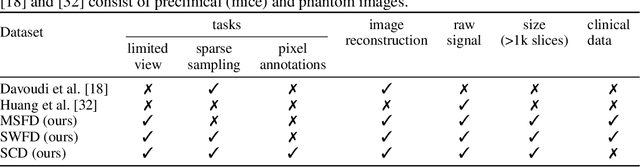
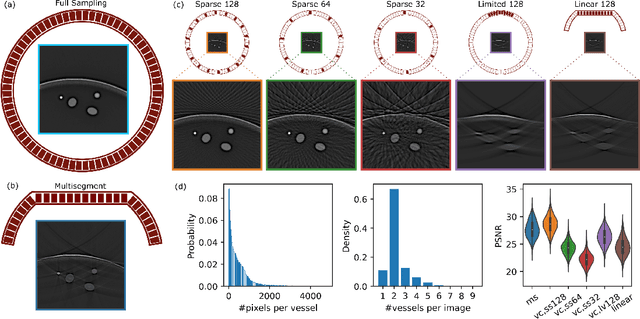
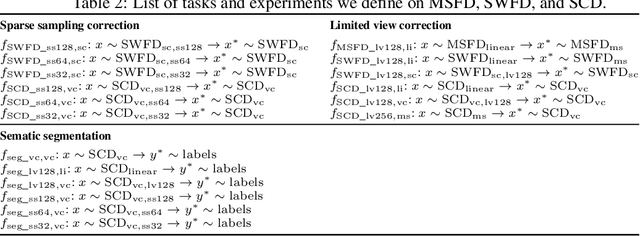
Abstract:Optoacoustic (OA) imaging is based on excitation of biological tissues with nanosecond-duration laser pulses followed by subsequent detection of ultrasound waves generated via light-absorption-mediated thermoelastic expansion. OA imaging features a powerful combination between rich optical contrast and high resolution in deep tissues. This enabled the exploration of a number of attractive new applications both in clinical and laboratory settings. However, no standardized datasets generated with different types of experimental set-up and associated processing methods are available to facilitate advances in broader applications of OA in clinical settings. This complicates an objective comparison between new and established data processing methods, often leading to qualitative results and arbitrary interpretations of the data. In this paper, we provide both experimental and synthetic OA raw signals and reconstructed image domain datasets rendered with different experimental parameters and tomographic acquisition geometries. We further provide trained neural networks to tackle three important challenges related to OA image processing, namely accurate reconstruction under limited view tomographic conditions, removal of spatial undersampling artifacts and anatomical segmentation for improved image reconstruction. Specifically, we define 18 experiments corresponding to the aforementioned challenges as benchmarks to be used as a reference for the development of more advanced processing methods.
Learning Summary Statistics for Bayesian Inference with Autoencoders
Jan 28, 2022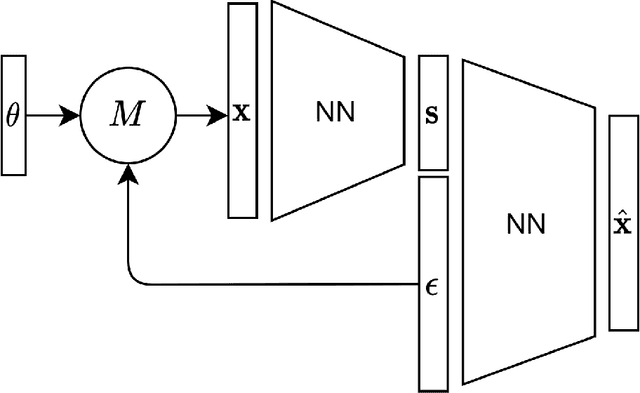
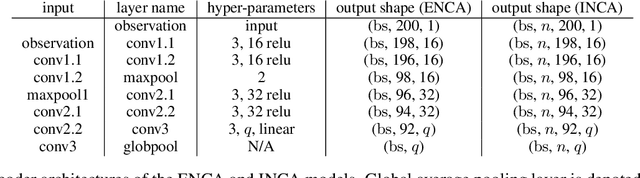
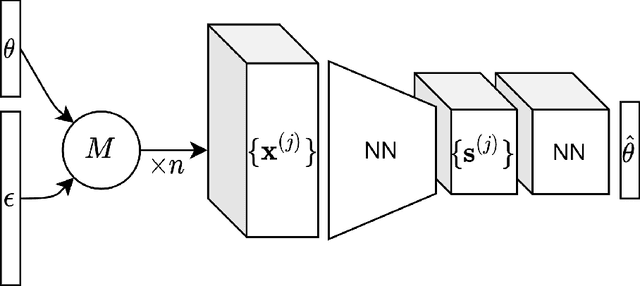
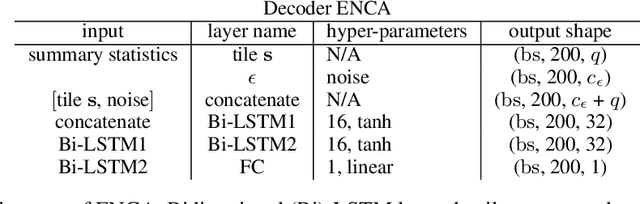
Abstract:For stochastic models with intractable likelihood functions, approximate Bayesian computation offers a way of approximating the true posterior through repeated comparisons of observations with simulated model outputs in terms of a small set of summary statistics. These statistics need to retain the information that is relevant for constraining the parameters but cancel out the noise. They can thus be seen as thermodynamic state variables, for general stochastic models. For many scientific applications, we need strictly more summary statistics than model parameters to reach a satisfactory approximation of the posterior. Therefore, we propose to use the inner dimension of deep neural network based Autoencoders as summary statistics. To create an incentive for the encoder to encode all the parameter-related information but not the noise, we give the decoder access to explicit or implicit information on the noise that has been used to generate the training data. We validate the approach empirically on two types of stochastic models.
Probabilistic modeling of lake surface water temperature using a Bayesian spatio-temporal graph convolutional neural network
Sep 27, 2021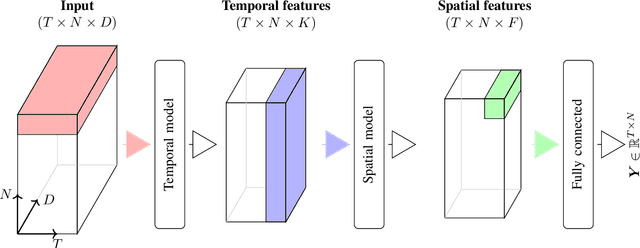
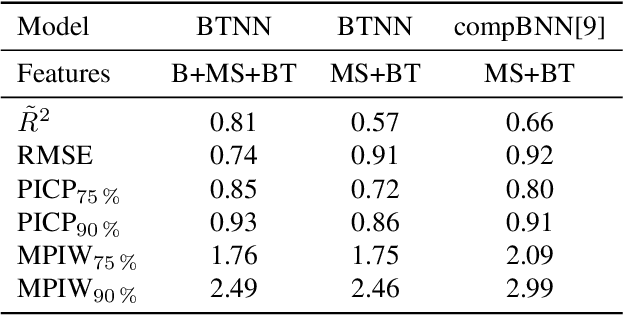
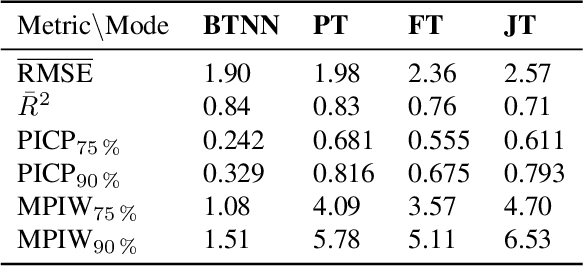
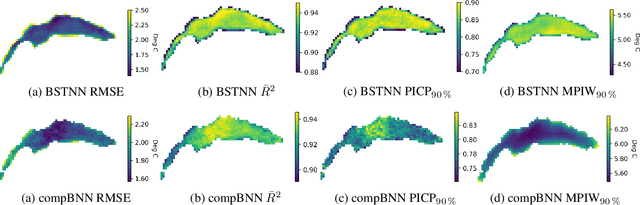
Abstract:Accurate lake temperature estimation is essential for numerous problems tackled in both hydrological and ecological domains. Nowadays physical models are developed to estimate lake dynamics; however, computations needed for accurate estimation of lake surface temperature can get prohibitively expensive. We propose to aggregate simulations of lake temperature at a certain depth together with a range of meteorological features to probabilistically estimate lake surface temperature. Accordingly, we introduce a spatio-temporal neural network that combines Bayesian recurrent neural networks and Bayesian graph convolutional neural networks. This work demonstrates that the proposed graphical model can deliver homogeneously good performance covering the whole lake surface despite having sparse training data available. Quantitative results are compared with a state-of-the-art Bayesian deep learning method. Code for the developed architectural layers, as well as demo scripts, are available on https://renkulab.io/projects/das/bstnn.
Delineating Bone Surfaces in B-Mode Images Constrained by Physics of Ultrasound Propagation
Jan 07, 2020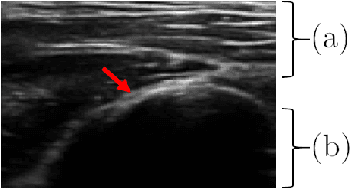
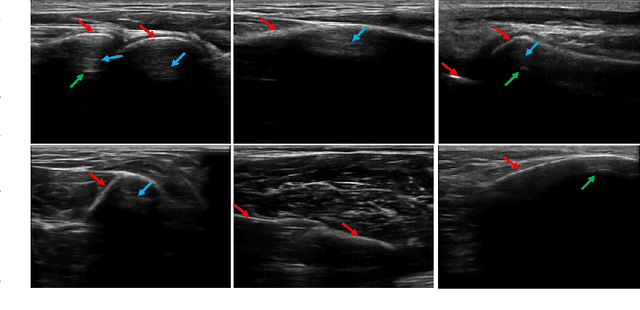
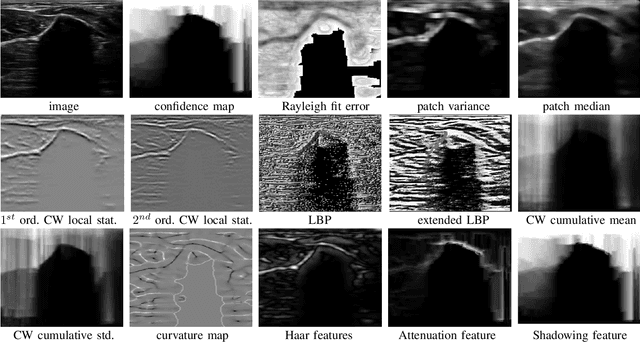
Abstract:Bone surface delineation in ultrasound is of interest due to its potential in diagnosis, surgical planning, and post-operative follow-up in orthopedics, as well as the potential of using bones as anatomical landmarks in surgical navigation. We herein propose a method to encode the physics of ultrasound propagation into a factor graph formulation for the purpose of bone surface delineation. In this graph structure, unary node potentials encode the local likelihood for being a soft tissue or acoustic-shadow (behind bone surface) region, both learned through image descriptors. Pair-wise edge potentials encode ultrasound propagation constraints of bone surfaces given their large acoustic-impedance difference. We evaluate the proposed method in comparison with four earlier approaches, on in-vivo ultrasound images collected from dorsal and volar views of the forearm. The proposed method achieves an average root-mean-square error and symmetric Hausdorff distance of 0.28mm and 1.78mm, respectively. It detects 99.9% of the annotated bone surfaces with a mean scanline error (distance to annotations) of 0.39mm.
Active Learning for Segmentation Based on Bayesian Sample Queries
Dec 22, 2019

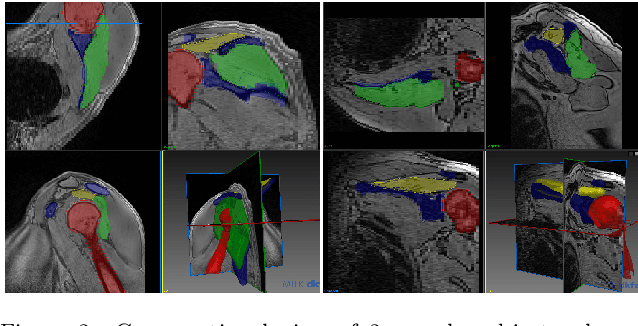
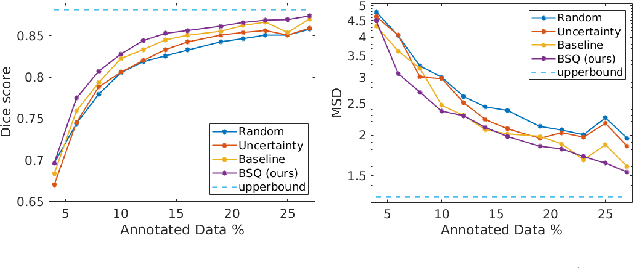
Abstract:Segmentation of anatomical structures is a fundamental image analysis task for many applications in the medical field. Deep learning methods have been shown to perform well, but for this purpose large numbers of manual annotations are needed in the first place, which necessitate prohibitive levels of resources that are often unavailable. In an active learning framework of selecting informed samples for manual labeling, expert clinician time for manual annotation can be optimally utilized, enabling the establishment of large labeled datasets for machine learning. In this paper, we propose a novel method that combines representativeness with uncertainty in order to estimate ideal samples to be annotated, iteratively from a given dataset. Our novel representativeness metric is based on Bayesian sampling, by using information-maximizing autoencoders. We conduct experiments on a shoulder magnetic resonance imaging (MRI) dataset for the segmentation of four musculoskeletal tissue classes. Quantitative results show that the annotation of representative samples selected by our proposed querying method yields an improved segmentation performance at each active learning iteration, compared to a baseline method that also employs uncertainty and representativeness metrics. For instance, with only 10% of the dataset annotated, our method reaches within 5% of Dice score expected from the upper bound scenario of all the dataset given as annotated (an impractical scenario due to resource constraints), and this gap drops down to a mere 2% when less than a fifth of the dataset samples are annotated. Such active learning approach to selecting samples to annotate enables an optimal use of the expert clinician time, being often the bottleneck in realizing machine learning solutions in medicine.
Extending Pretrained Segmentation Networks with Additional Anatomical Structures
Nov 12, 2018
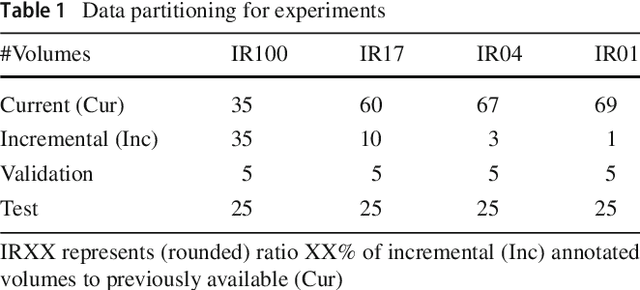

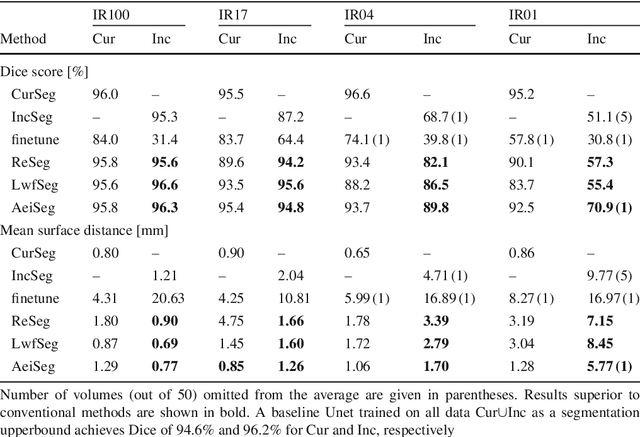
Abstract:Comprehensive surgical planning require complex patient-specific anatomical models. For instance, functional muskuloskeletal simulations necessitate all relevant structures to be segmented, which could be performed in real-time using deep neural networks given sufficient annotated samples. Such large datasets of multiple structure annotations are costly to procure and are often unavailable in practice. Nevertheless, annotations from different studies and centers can be readily available, or become available in the future in an incremental fashion. We propose a class-incremental segmentation framework for extending a deep network trained for some anatomical structure to yet another structure using a small incremental annotation set. Through distilling knowledge from the current state of the framework, we bypass the need for a full retraining. This is a meta-method to extend any choice of desired deep segmentation network with only a minor addition per structure, which makes it suitable for lifelong class-incremental learning and applicable also for future deep neural network architectures. We evaluated our methods on a public knee dataset of 100 MR volumes. Through varying amount of incremental annotation ratios, we show how our proposed method can retain the previous anatomical structure segmentation performance superior to the conventional finetuning approach. In addition, our framework inherently exploits transferable knowledge from previously trained structures to incremental tasks, demonstrated by superior results compared to non-incremental training. With the presented method, new anatomical structures can be learned without catastrophic forgetting of older structures and without extensive increase of memory and complexity.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge