Philipp Fuernstahl
MIXPINN: Mixed-Material Simulations by Physics-Informed Neural Network
Mar 17, 2025Abstract:Simulating the complex interactions between soft tissues and rigid anatomy is critical for applications in surgical training, planning, and robotic-assisted interventions. Traditional Finite Element Method (FEM)-based simulations, while accurate, are computationally expensive and impractical for real-time scenarios. Learning-based approaches have shown promise in accelerating predictions but have fallen short in modeling soft-rigid interactions effectively. We introduce MIXPINN, a physics-informed Graph Neural Network (GNN) framework for mixed-material simulations, explicitly capturing soft-rigid interactions using graph-based augmentations. Our approach integrates Virtual Nodes (VNs) and Virtual Edges (VEs) to enhance rigid body constraint satisfaction while preserving computational efficiency. By leveraging a graph-based representation of biomechanical structures, MIXPINN learns high-fidelity deformations from FEM-generated data and achieves real-time inference with sub-millimeter accuracy. We validate our method in a realistic clinical scenario, demonstrating superior performance compared to baseline GNN models and traditional FEM methods. Our results show that MIXPINN reduces computational cost by an order of magnitude while maintaining high physical accuracy, making it a viable solution for real-time surgical simulation and robotic-assisted procedures.
Safe Deep RL for Intraoperative Planning of Pedicle Screw Placement
May 10, 2023



Abstract:Spinal fusion surgery requires highly accurate implantation of pedicle screw implants, which must be conducted in critical proximity to vital structures with a limited view of anatomy. Robotic surgery systems have been proposed to improve placement accuracy, however, state-of-the-art systems suffer from the limitations of open-loop approaches, as they follow traditional concepts of preoperative planning and intraoperative registration, without real-time recalculation of the surgical plan. In this paper, we propose an intraoperative planning approach for robotic spine surgery that leverages real-time observation for drill path planning based on Safe Deep Reinforcement Learning (DRL). The main contributions of our method are (1) the capability to guarantee safe actions by introducing an uncertainty-aware distance-based safety filter; and (2) the ability to compensate for incomplete intraoperative anatomical information, by encoding a-priori knowledge about anatomical structures with a network pre-trained on high-fidelity anatomical models. Planning quality was assessed by quantitative comparison with the gold standard (GS) drill planning. In experiments with 5 models derived from real magnetic resonance imaging (MRI) data, our approach was capable of achieving 90% bone penetration with respect to the GS while satisfying safety requirements, even under observation and motion uncertainty. To the best of our knowledge, our approach is the first safe DRL approach focusing on orthopedic surgeries.
Active Learning for Segmentation Based on Bayesian Sample Queries
Dec 22, 2019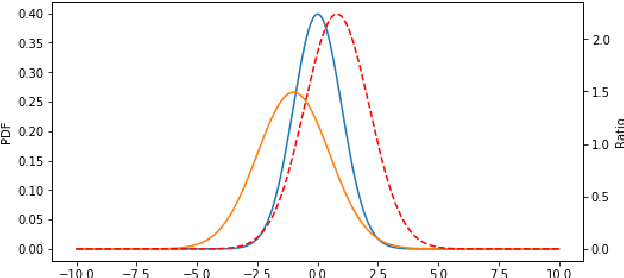

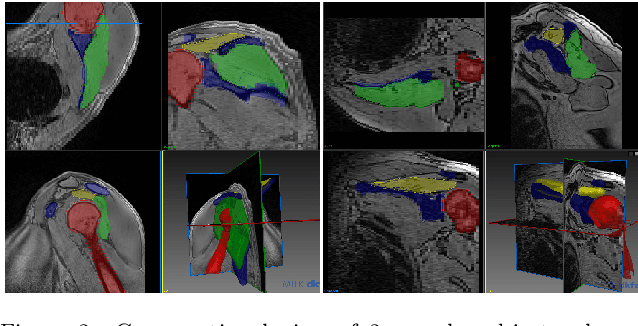
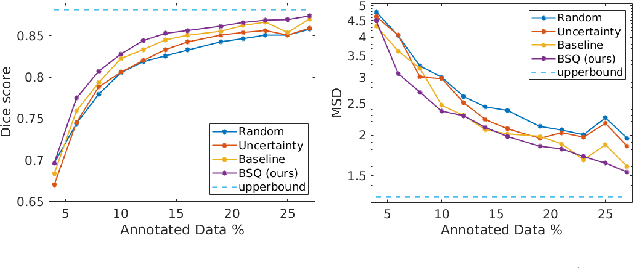
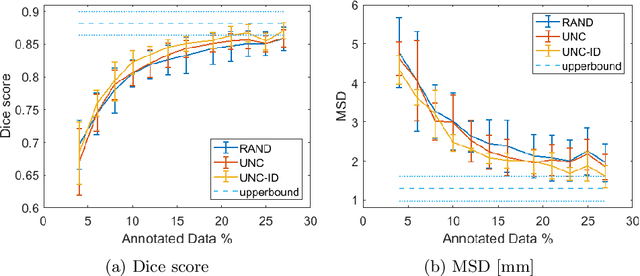
Abstract:Segmentation of anatomical structures is a fundamental image analysis task for many applications in the medical field. Deep learning methods have been shown to perform well, but for this purpose large numbers of manual annotations are needed in the first place, which necessitate prohibitive levels of resources that are often unavailable. In an active learning framework of selecting informed samples for manual labeling, expert clinician time for manual annotation can be optimally utilized, enabling the establishment of large labeled datasets for machine learning. In this paper, we propose a novel method that combines representativeness with uncertainty in order to estimate ideal samples to be annotated, iteratively from a given dataset. Our novel representativeness metric is based on Bayesian sampling, by using information-maximizing autoencoders. We conduct experiments on a shoulder magnetic resonance imaging (MRI) dataset for the segmentation of four musculoskeletal tissue classes. Quantitative results show that the annotation of representative samples selected by our proposed querying method yields an improved segmentation performance at each active learning iteration, compared to a baseline method that also employs uncertainty and representativeness metrics. For instance, with only 10% of the dataset annotated, our method reaches within 5% of Dice score expected from the upper bound scenario of all the dataset given as annotated (an impractical scenario due to resource constraints), and this gap drops down to a mere 2% when less than a fifth of the dataset samples are annotated. Such active learning approach to selecting samples to annotate enables an optimal use of the expert clinician time, being often the bottleneck in realizing machine learning solutions in medicine.
Active Learning for Segmentation by Optimizing Content Information for Maximal Entropy
Jul 18, 2018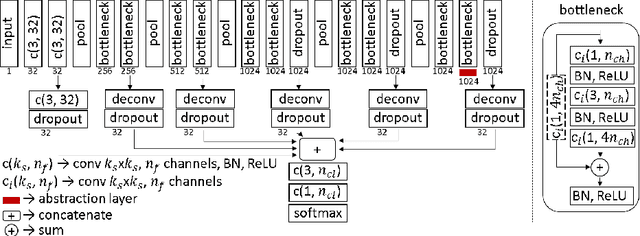

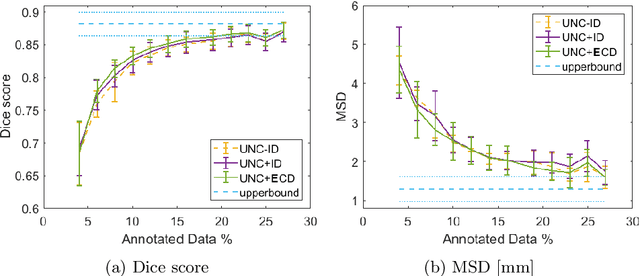
Abstract:Segmentation is essential for medical image analysis tasks such as intervention planning, therapy guidance, diagnosis, treatment decisions. Deep learning is becoming increasingly prominent for segmentation, where the lack of annotations, however, often becomes the main limitation. Due to privacy concerns and ethical considerations, most medical datasets are created, curated, and allow access only locally. Furthermore, current deep learning methods are often suboptimal in translating anatomical knowledge between different medical imaging modalities. Active learning can be used to select an informed set of image samples to request for manual annotation, in order to best utilize the limited annotation time of clinical experts for optimal outcomes, which we focus on in this work. Our contributions herein are two fold: (1) we enforce domain-representativeness of selected samples using a proposed penalization scheme to maximize information at the network abstraction layer, and (2) we propose a Borda-count based sample querying scheme for selecting samples for segmentation. Comparative experiments with baseline approaches show that the samples queried with our proposed method, where both above contributions are combined, result in significantly improved segmentation performance for this active learning task.
Learn the new, keep the old: Extending pretrained models with new anatomy and images
Jun 01, 2018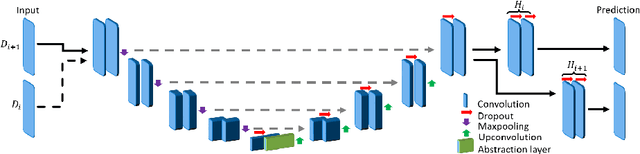
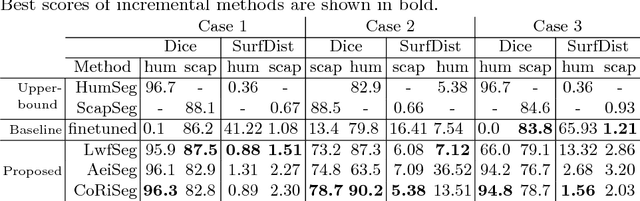
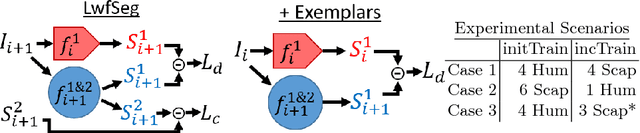
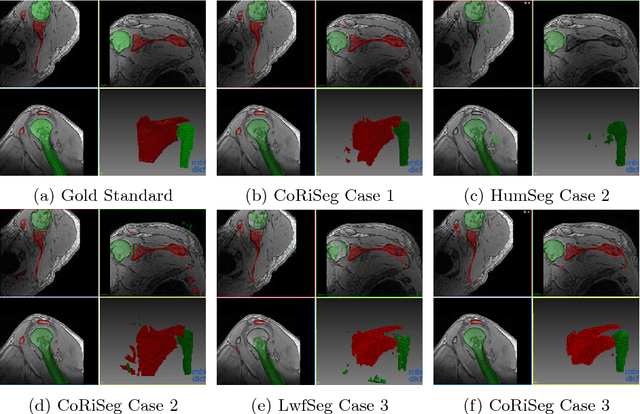
Abstract:Deep learning has been widely accepted as a promising solution for medical image segmentation, given a sufficiently large representative dataset of images with corresponding annotations. With ever increasing amounts of annotated medical datasets, it is infeasible to train a learning method always with all data from scratch. This is also doomed to hit computational limits, e.g., memory or runtime feasible for training. Incremental learning can be a potential solution, where new information (images or anatomy) is introduced iteratively. Nevertheless, for the preservation of the collective information, it is essential to keep some "important" (i.e. representative) images and annotations from the past, while adding new information. In this paper, we introduce a framework for applying incremental learning for segmentation and propose novel methods for selecting representative data therein. We comparatively evaluate our methods in different scenarios using MR images and validate the increased learning capacity with using our methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge