Zixuan Peng
Efficient Speech Emotion Recognition Using Multi-Scale CNN and Attention
Jun 08, 2021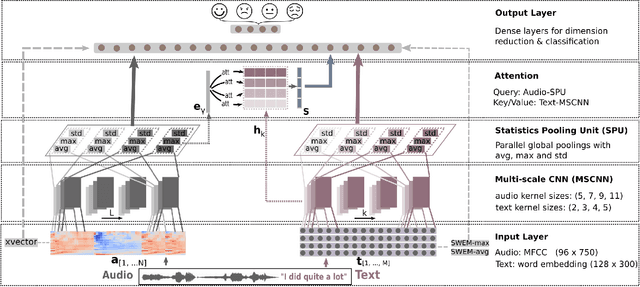
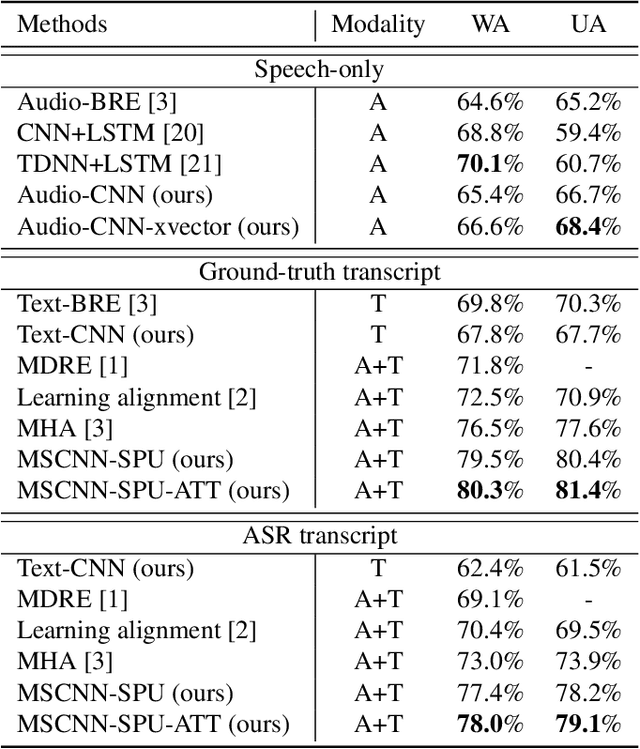

Abstract:Emotion recognition from speech is a challenging task. Re-cent advances in deep learning have led bi-directional recur-rent neural network (Bi-RNN) and attention mechanism as astandard method for speech emotion recognition, extractingand attending multi-modal features - audio and text, and thenfusing them for downstream emotion classification tasks. Inthis paper, we propose a simple yet efficient neural networkarchitecture to exploit both acoustic and lexical informationfrom speech. The proposed framework using multi-scale con-volutional layers (MSCNN) to obtain both audio and text hid-den representations. Then, a statistical pooling unit (SPU)is used to further extract the features in each modality. Be-sides, an attention module can be built on top of the MSCNN-SPU (audio) and MSCNN (text) to further improve the perfor-mance. Extensive experiments show that the proposed modeloutperforms previous state-of-the-art methods on IEMOCAPdataset with four emotion categories (i.e., angry, happy, sadand neutral) in both weighted accuracy (WA) and unweightedaccuracy (UA), with an improvement of 5.0% and 5.2% respectively under the ASR setting.
* First two authors contributed equally.Accepted by ICASSP 2021
Active Learning for Segmentation Based on Bayesian Sample Queries
Dec 22, 2019

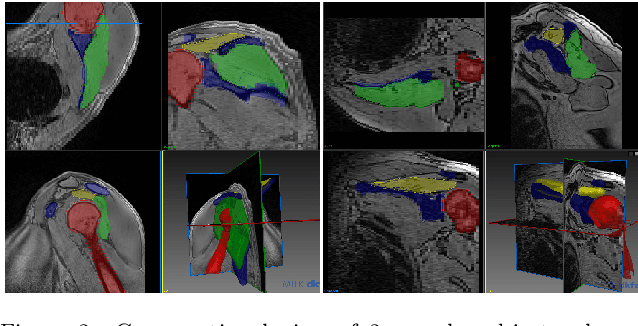
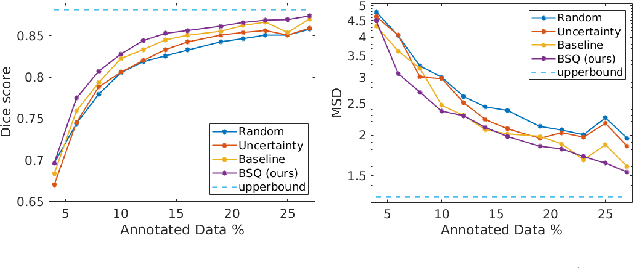
Abstract:Segmentation of anatomical structures is a fundamental image analysis task for many applications in the medical field. Deep learning methods have been shown to perform well, but for this purpose large numbers of manual annotations are needed in the first place, which necessitate prohibitive levels of resources that are often unavailable. In an active learning framework of selecting informed samples for manual labeling, expert clinician time for manual annotation can be optimally utilized, enabling the establishment of large labeled datasets for machine learning. In this paper, we propose a novel method that combines representativeness with uncertainty in order to estimate ideal samples to be annotated, iteratively from a given dataset. Our novel representativeness metric is based on Bayesian sampling, by using information-maximizing autoencoders. We conduct experiments on a shoulder magnetic resonance imaging (MRI) dataset for the segmentation of four musculoskeletal tissue classes. Quantitative results show that the annotation of representative samples selected by our proposed querying method yields an improved segmentation performance at each active learning iteration, compared to a baseline method that also employs uncertainty and representativeness metrics. For instance, with only 10% of the dataset annotated, our method reaches within 5% of Dice score expected from the upper bound scenario of all the dataset given as annotated (an impractical scenario due to resource constraints), and this gap drops down to a mere 2% when less than a fifth of the dataset samples are annotated. Such active learning approach to selecting samples to annotate enables an optimal use of the expert clinician time, being often the bottleneck in realizing machine learning solutions in medicine.
Active Learning for Segmentation by Optimizing Content Information for Maximal Entropy
Jul 18, 2018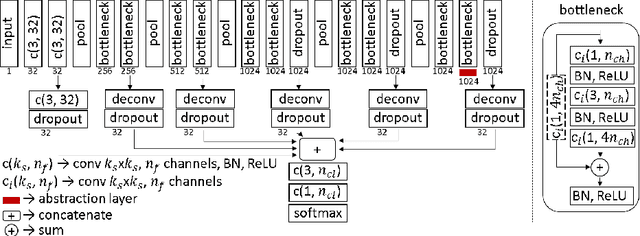

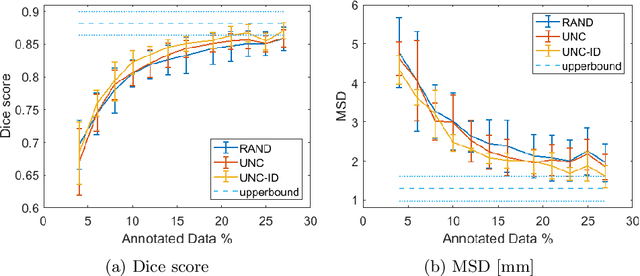
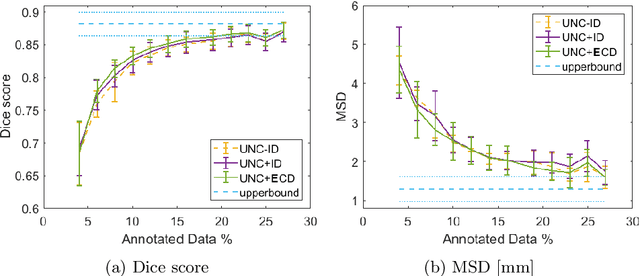
Abstract:Segmentation is essential for medical image analysis tasks such as intervention planning, therapy guidance, diagnosis, treatment decisions. Deep learning is becoming increasingly prominent for segmentation, where the lack of annotations, however, often becomes the main limitation. Due to privacy concerns and ethical considerations, most medical datasets are created, curated, and allow access only locally. Furthermore, current deep learning methods are often suboptimal in translating anatomical knowledge between different medical imaging modalities. Active learning can be used to select an informed set of image samples to request for manual annotation, in order to best utilize the limited annotation time of clinical experts for optimal outcomes, which we focus on in this work. Our contributions herein are two fold: (1) we enforce domain-representativeness of selected samples using a proposed penalization scheme to maximize information at the network abstraction layer, and (2) we propose a Borda-count based sample querying scheme for selecting samples for segmentation. Comparative experiments with baseline approaches show that the samples queried with our proposed method, where both above contributions are combined, result in significantly improved segmentation performance for this active learning task.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge