Orcun Goksel
ETH Zurich, Uppsala University
Uncertainty Estimation for Trust Attribution to Speed-of-Sound Reconstruction with Variational Networks
Apr 15, 2025Abstract:Speed-of-sound (SoS) is a biomechanical characteristic of tissue, and its imaging can provide a promising biomarker for diagnosis. Reconstructing SoS images from ultrasound acquisitions can be cast as a limited-angle computed-tomography problem, with Variational Networks being a promising model-based deep learning solution. Some acquired data frames may, however, get corrupted by noise due to, e.g., motion, lack of contact, and acoustic shadows, which in turn negatively affects the resulting SoS reconstructions. We propose to use the uncertainty in SoS reconstructions to attribute trust to each individual acquired frame. Given multiple acquisitions, we then use an uncertainty based automatic selection among these retrospectively, to improve diagnostic decisions. We investigate uncertainty estimation based on Monte Carlo Dropout and Bayesian Variational Inference. We assess our automatic frame selection method for differential diagnosis of breast cancer, distinguishing between benign fibroadenoma and malignant carcinoma. We evaluate 21 lesions classified as BI-RADS~4, which represents suspicious cases for probable malignancy. The most trustworthy frame among four acquisitions of each lesion was identified using uncertainty based criteria. Selecting a frame informed by uncertainty achieved an area under curve of 76% and 80% for Monte Carlo Dropout and Bayesian Variational Inference, respectively, superior to any uncertainty-uninformed baselines with the best one achieving 64%. A novel use of uncertainty estimation is proposed for selecting one of multiple data acquisitions for further processing and decision making.
Image-Based Metrics in Ultrasound for Estimation of Global Speed-of-Sound
Mar 18, 2025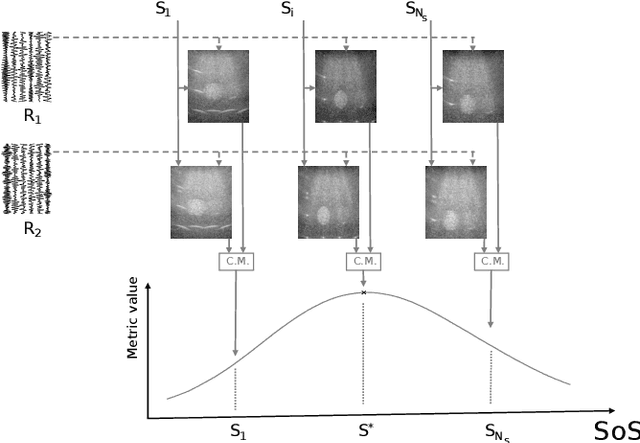
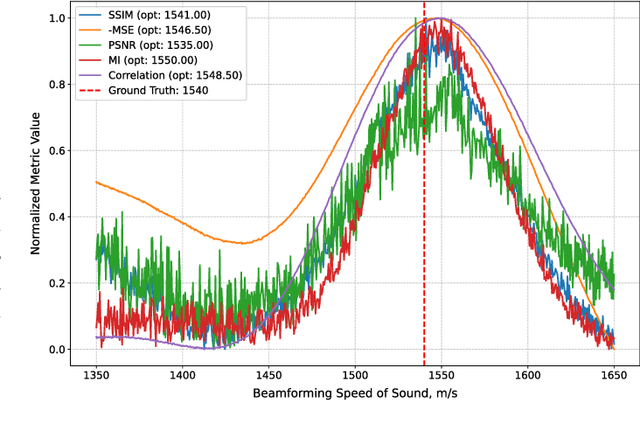
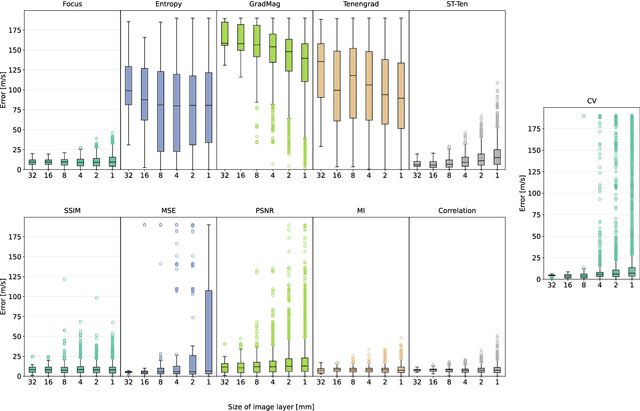
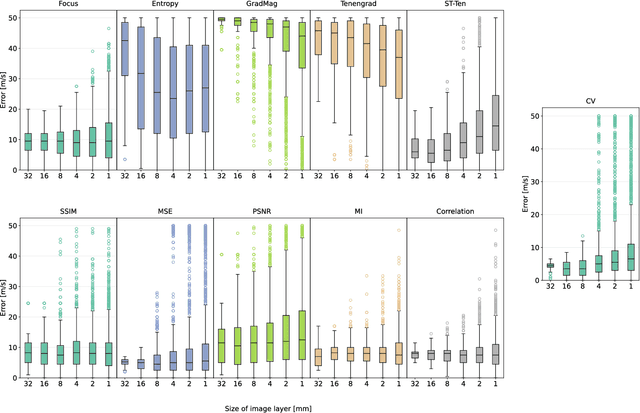
Abstract:Accurate speed-of-sound (SoS) estimation is crucial for ultrasound image formation, yet conventional systems often rely on an assumed value for imaging. While several methods exist for SoS estimation, they typically depend on complex physical models of acoustic propagation. We propose to leverage conventional image analysis techniques and metrics, as a novel and simple approach to estimate tissue SoS. We study eleven metrics in three categories for assessing image quality, image similarity and multi-frame variation, by testing them in numerical simulations and phantom experiments. Among single-frame image quality metrics, conventional Focus and our proposed Smoothed Threshold Tenengrad metrics achieved satisfactory accuracy, however only when applied to compounded images. Image quality metrics were largely surpassed by various image comparison metrics, which exhibited errors consistently under 8 m/s even applied to a single pair of images. Particularly, Mean Square Error is a computationally efficient alternative for global estimation. Mutual Information and Correlation are found to be robust to processing small image segments, making them suitable, e.g., for multi-layer SoS estimation. The above metrics do not require access to raw channel data as they can operate on post-beamformed data, and in the case of image quality metrics they can operate on B-mode images, given that the beamforming SoS can be controlled for beamforming using a multitude of values. These image analysis based SoS estimation methods offer a computationally efficient and data-accessible alternative to conventional physics-based methods, with potential extensions to layered or local SoS imaging.
FGGP: Fixed-Rate Gradient-First Gradual Pruning
Nov 08, 2024Abstract:In recent years, the increasing size of deep learning models and their growing demand for computational resources have drawn significant attention to the practice of pruning neural networks, while aiming to preserve their accuracy. In unstructured gradual pruning, which sparsifies a network by gradually removing individual network parameters until a targeted network sparsity is reached, recent works show that both gradient and weight magnitudes should be considered. In this work, we show that such mechanism, e.g., the order of prioritization and selection criteria, is essential. We introduce a gradient-first magnitude-next strategy for choosing the parameters to prune, and show that a fixed-rate subselection criterion between these steps works better, in contrast to the annealing approach in the literature. We validate this on CIFAR-10 dataset, with multiple randomized initializations on both VGG-19 and ResNet-50 network backbones, for pruning targets of 90, 95, and 98% sparsity and for both initially dense and 50% sparse networks. Our proposed fixed-rate gradient-first gradual pruning (FGGP) approach outperforms its state-of-the-art alternatives in most of the above experimental settings, even occasionally surpassing the upperbound of corresponding dense network results, and having the highest ranking across the considered experimental settings.
Joint semi-supervised and contrastive learning enables zero-shot domain-adaptation and multi-domain segmentation
May 08, 2024



Abstract:Despite their effectiveness, current deep learning models face challenges with images coming from different domains with varying appearance and content. We introduce SegCLR, a versatile framework designed to segment volumetric images across different domains, employing supervised and contrastive learning simultaneously to effectively learn from both labeled and unlabeled data. We demonstrate the superior performance of SegCLR through a comprehensive evaluation involving three diverse clinical datasets of retinal fluid segmentation in 3D Optical Coherence Tomography (OCT), various network configurations, and verification across 10 different network initializations. In an unsupervised domain adaptation context, SegCLR achieves results on par with a supervised upper-bound model trained on the intended target domain. Notably, we discover that the segmentation performance of SegCLR framework is marginally impacted by the abundance of unlabeled data from the target domain, thereby we also propose an effective zero-shot domain adaptation extension of SegCLR, eliminating the need for any target domain information. This shows that our proposed addition of contrastive loss in standard supervised training for segmentation leads to superior models, inherently more generalizable to both in- and out-of-domain test data. We additionally propose a pragmatic solution for SegCLR deployment in realistic scenarios with multiple domains containing labeled data. Accordingly, our framework pushes the boundaries of deep-learning based segmentation in multi-domain applications, regardless of data availability - labeled, unlabeled, or nonexistent.
Learning the Imaging Model of Speed-of-Sound Reconstruction via a Convolutional Formulation
Sep 01, 2023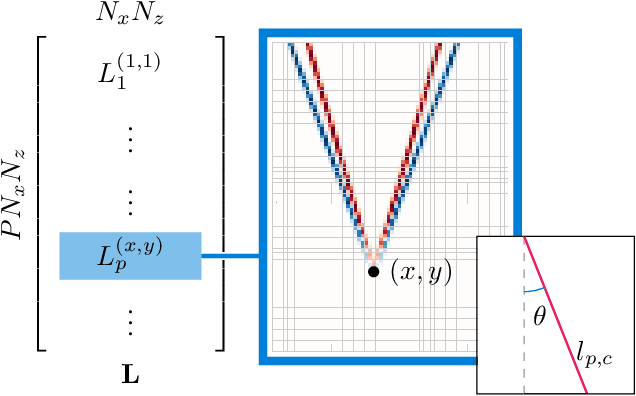
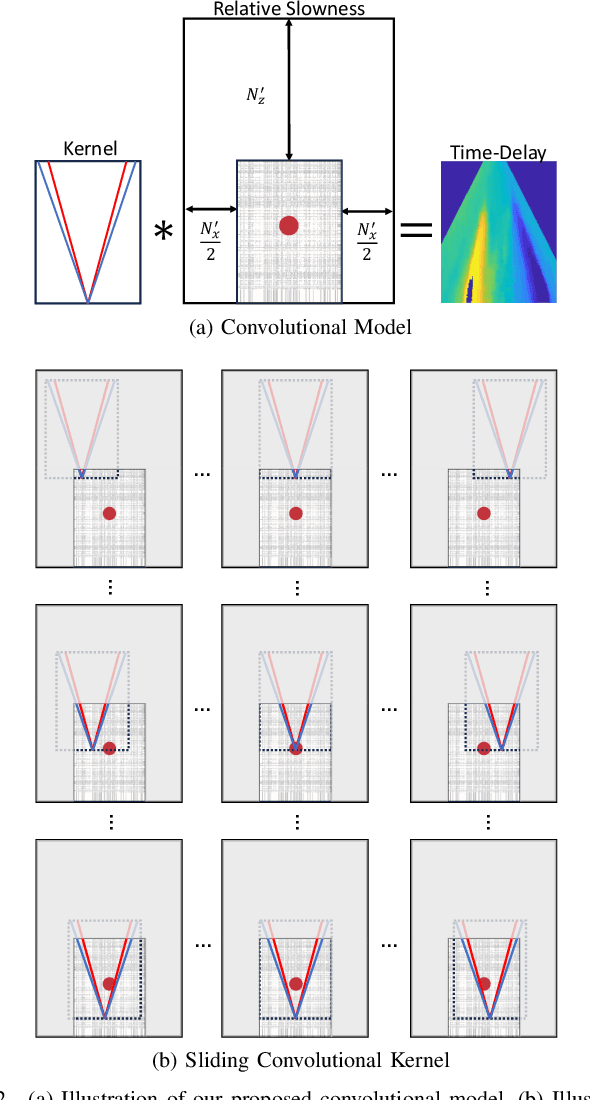
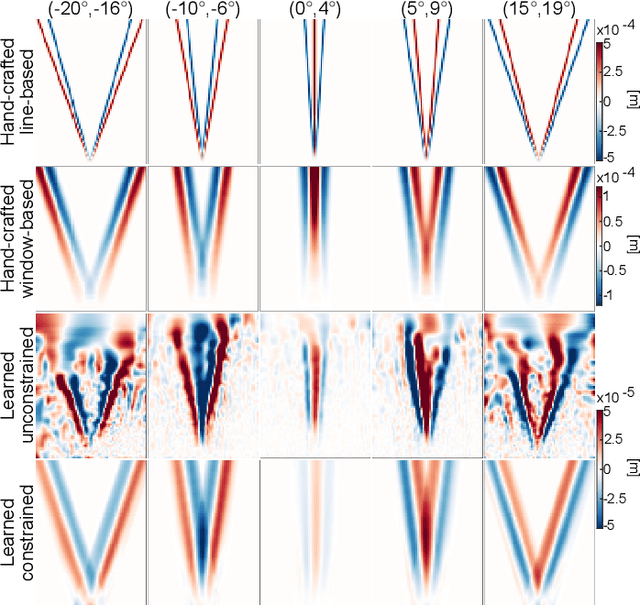
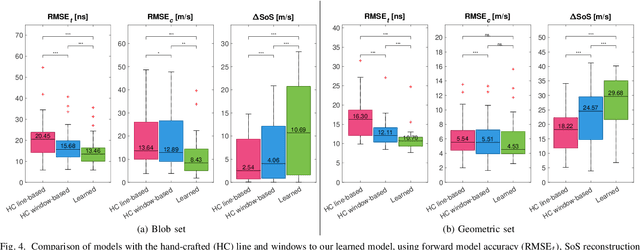
Abstract:Speed-of-sound (SoS) is an emerging ultrasound contrast modality, where pulse-echo techniques using conventional transducers offer multiple benefits. For estimating tissue SoS distributions, spatial domain reconstruction from relative speckle shifts between different beamforming sequences is a promising approach. This operates based on a forward model that relates the sought local values of SoS to observed speckle shifts, for which the associated image reconstruction inverse problem is solved. The reconstruction accuracy thus highly depends on the hand-crafted forward imaging model. In this work, we propose to learn the SoS imaging model based on data. We introduce a convolutional formulation of the pulse-echo SoS imaging problem such that the entire field-of-view requires a single unified kernel, the learning of which is then tractable and robust. We present least-squares estimation of such convolutional kernel, which can further be constrained and regularized for numerical stability. In experiments, we show that a forward model learned from k-Wave simulations improves the median contrast of SoS reconstructions by 63%, compared to a conventional hand-crafted line-based wave-path model. This simulation-learned model generalizes successfully to acquired phantom data, nearly doubling the SoS contrast compared to the conventional hand-crafted alternative. We demonstrate equipment-specific and small-data regime feasibility by learning a forward model from a single phantom image, where our learned model quadruples the SoS contrast compared to the conventional hand-crafted model. On in-vivo data, the simulation- and phantom-learned models respectively exhibit impressive 7 and 10 folds contrast improvements over the conventional model.
Robust Imaging of Speed-of-Sound Using Virtual Source Transmission
Mar 20, 2023Abstract:Speed-of-sound (SoS) is a novel imaging biomarker for assessing biomechanical characteristics of soft tissues. SoS imaging in pulse-echo mode using conventional ultrasound systems with hand-held transducers has the potential to enable new clinical uses. Recent work demonstrated diverging waves from single-element (SE) transmits to outperform plane-wave sequences. However, single-element transmits have severely limited power and hence produce low signal-to-noise ratio (SNR) in echo data. We herein propose Walsh-Hadamard (WH) coded and virtual-source (VS) transmit sequences for improved SNR in SoS imaging. We additionally present an iterative method of estimating beamforming SoS in the medium, which otherwise confound SoS reconstructions due to beamforming inaccuracies in the images used for reconstruction. Through numerical simulations, phantom experiments, and in-vivo imaging data, we show that WH is not robust against motion, which is often unavoidable in clinical imaging scenarios. Our proposed virtual-source sequence is shown to provide the highest SoS reconstruction performance, especially robust to motion artifacts. In phantom experiments, despite having a comparable SoS root-mean-square-error (RMSE) of 17.5 to 18.0 m/s at rest, with a minor axial probe motion of ~0.67 mm/s the RMSE for SE, WH, and VS already deteriorate to 20.2, 105.4, 19.0 m/s, respectively; showing that WH produces unacceptable results, not robust to motion. In the clinical data, the high SNR and motion-resilience of VS sequence is seen to yield superior contrast compared to SE and WH sequences.
Unpaired Translation from Semantic Label Maps to Images by Leveraging Domain-Specific Simulations
Feb 21, 2023Abstract:Photorealistic image generation from simulated label maps are necessitated in several contexts, such as for medical training in virtual reality. With conventional deep learning methods, this task requires images that are paired with semantic annotations, which typically are unavailable. We introduce a contrastive learning framework for generating photorealistic images from simulated label maps, by learning from unpaired sets of both. Due to potentially large scene differences between real images and label maps, existing unpaired image translation methods lead to artifacts of scene modification in synthesized images. We utilize simulated images as surrogate targets for a contrastive loss, while ensuring consistency by utilizing features from a reverse translation network. Our method enables bidirectional label-image translations, which is demonstrated in a variety of scenarios and datasets, including laparoscopy, ultrasound, and driving scenes. By comparing with state-of-the-art unpaired translation methods, our proposed method is shown to generate realistic and scene-accurate translations.
Multi-scale Feature Alignment for Continual Learning of Unlabeled Domains
Feb 02, 2023Abstract:Methods for unsupervised domain adaptation (UDA) help to improve the performance of deep neural networks on unseen domains without any labeled data. Especially in medical disciplines such as histopathology, this is crucial since large datasets with detailed annotations are scarce. While the majority of existing UDA methods focus on the adaptation from a labeled source to a single unlabeled target domain, many real-world applications with a long life cycle involve more than one target domain. Thus, the ability to sequentially adapt to multiple target domains becomes essential. In settings where the data from previously seen domains cannot be stored, e.g., due to data protection regulations, the above becomes a challenging continual learning problem. To this end, we propose to use generative feature-driven image replay in conjunction with a dual-purpose discriminator that not only enables the generation of images with realistic features for replay, but also promotes feature alignment during domain adaptation. We evaluate our approach extensively on a sequence of three histopathological datasets for tissue-type classification, achieving state-of-the-art results. We present detailed ablation experiments studying our proposed method components and demonstrate a possible use-case of our continual UDA method for an unsupervised patch-based segmentation task given high-resolution tissue images.
Weakly Supervised Joint Whole-Slide Segmentation and Classification in Prostate Cancer
Jan 07, 2023Abstract:The segmentation and automatic identification of histological regions of diagnostic interest offer a valuable aid to pathologists. However, segmentation methods are hampered by the difficulty of obtaining pixel-level annotations, which are tedious and expensive to obtain for Whole-Slide images (WSI). To remedy this, weakly supervised methods have been developed to exploit the annotations directly available at the image level. However, to our knowledge, none of these techniques is adapted to deal with WSIs. In this paper, we propose WholeSIGHT, a weakly-supervised method, to simultaneously segment and classify WSIs of arbitrary shapes and sizes. Formally, WholeSIGHT first constructs a tissue-graph representation of the WSI, where the nodes and edges depict tissue regions and their interactions, respectively. During training, a graph classification head classifies the WSI and produces node-level pseudo labels via post-hoc feature attribution. These pseudo labels are then used to train a node classification head for WSI segmentation. During testing, both heads simultaneously render class prediction and segmentation for an input WSI. We evaluated WholeSIGHT on three public prostate cancer WSI datasets. Our method achieved state-of-the-art weakly-supervised segmentation performance on all datasets while resulting in better or comparable classification with respect to state-of-the-art weakly-supervised WSI classification methods. Additionally, we quantify the generalization capability of our method in terms of segmentation and classification performance, uncertainty estimation, and model calibration.
Generative appearance replay for continual unsupervised domain adaptation
Jan 03, 2023



Abstract:Deep learning models can achieve high accuracy when trained on large amounts of labeled data. However, real-world scenarios often involve several challenges: Training data may become available in installments, may originate from multiple different domains, and may not contain labels for training. Certain settings, for instance medical applications, often involve further restrictions that prohibit retention of previously seen data due to privacy regulations. In this work, to address such challenges, we study unsupervised segmentation in continual learning scenarios that involve domain shift. To that end, we introduce GarDA (Generative Appearance Replay for continual Domain Adaptation), a generative-replay based approach that can adapt a segmentation model sequentially to new domains with unlabeled data. In contrast to single-step unsupervised domain adaptation (UDA), continual adaptation to a sequence of domains enables leveraging and consolidation of information from multiple domains. Unlike previous approaches in incremental UDA, our method does not require access to previously seen data, making it applicable in many practical scenarios. We evaluate GarDA on two datasets with different organs and modalities, where it substantially outperforms existing techniques.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge