Alvaro Gomariz
Characterizing the Interpretability of Attention Maps in Digital Pathology
Jul 02, 2024



Abstract:Interpreting machine learning model decisions is crucial for high-risk applications like healthcare. In digital pathology, large whole slide images (WSIs) are decomposed into smaller tiles and tile-derived features are processed by attention-based multiple instance learning (ABMIL) models to predict WSI-level labels. These networks generate tile-specific attention weights, which can be visualized as attention maps for interpretability. However, a standardized evaluation framework for these maps is lacking, questioning their reliability and ability to detect spurious correlations that can mislead models. We herein propose a framework to assess the ability of attention networks to attend to relevant features in digital pathology by creating artificial model confounders and using dedicated interpretability metrics. Models are trained and evaluated on data with tile modifications correlated with WSI labels, enabling the analysis of model sensitivity to artificial confounders and the accuracy of attention maps in highlighting them. Confounders are introduced either through synthetic tile modifications or through tile ablations based on their specific image-based features, with the latter being used to assess more clinically relevant scenarios. We also analyze the impact of varying confounder quantities at both the tile and WSI levels. Our results show that ABMIL models perform as desired within our framework. While attention maps generally highlight relevant regions, their robustness is affected by the type and number of confounders. Our versatile framework has the potential to be used in the evaluation of various methods and the exploration of image-based features driving model predictions, which could aid in biomarker discovery.
Joint semi-supervised and contrastive learning enables zero-shot domain-adaptation and multi-domain segmentation
May 08, 2024



Abstract:Despite their effectiveness, current deep learning models face challenges with images coming from different domains with varying appearance and content. We introduce SegCLR, a versatile framework designed to segment volumetric images across different domains, employing supervised and contrastive learning simultaneously to effectively learn from both labeled and unlabeled data. We demonstrate the superior performance of SegCLR through a comprehensive evaluation involving three diverse clinical datasets of retinal fluid segmentation in 3D Optical Coherence Tomography (OCT), various network configurations, and verification across 10 different network initializations. In an unsupervised domain adaptation context, SegCLR achieves results on par with a supervised upper-bound model trained on the intended target domain. Notably, we discover that the segmentation performance of SegCLR framework is marginally impacted by the abundance of unlabeled data from the target domain, thereby we also propose an effective zero-shot domain adaptation extension of SegCLR, eliminating the need for any target domain information. This shows that our proposed addition of contrastive loss in standard supervised training for segmentation leads to superior models, inherently more generalizable to both in- and out-of-domain test data. We additionally propose a pragmatic solution for SegCLR deployment in realistic scenarios with multiple domains containing labeled data. Accordingly, our framework pushes the boundaries of deep-learning based segmentation in multi-domain applications, regardless of data availability - labeled, unlabeled, or nonexistent.
Integrating multiscale topology in digital pathology with pyramidal graph convolutional networks
Mar 22, 2024Abstract:Graph convolutional networks (GCNs) have emerged as a powerful alternative to multiple instance learning with convolutional neural networks in digital pathology, offering superior handling of structural information across various spatial ranges - a crucial aspect of learning from gigapixel H&E-stained whole slide images (WSI). However, graph message-passing algorithms often suffer from oversmoothing when aggregating a large neighborhood. Hence, effective modeling of multi-range interactions relies on the careful construction of the graph. Our proposed multi-scale GCN (MS-GCN) tackles this issue by leveraging information across multiple magnification levels in WSIs. MS-GCN enables the simultaneous modeling of long-range structural dependencies at lower magnifications and high-resolution cellular details at higher magnifications, akin to analysis pipelines usually conducted by pathologists. The architecture's unique configuration allows for the concurrent modeling of structural patterns at lower magnifications and detailed cellular features at higher ones, while also quantifying the contribution of each magnification level to the prediction. Through testing on different datasets, MS-GCN demonstrates superior performance over existing single-magnification GCN methods. The enhancement in performance and interpretability afforded by our method holds promise for advancing computational pathology models, especially in tasks requiring extensive spatial context.
Aggregation Model Hyperparameters Matter in Digital Pathology
Nov 29, 2023Abstract:Digital pathology has significantly advanced disease detection and pathologist efficiency through the analysis of gigapixel whole-slide images (WSI). In this process, WSIs are first divided into patches, for which a feature extractor model is applied to obtain feature vectors, which are subsequently processed by an aggregation model to predict the respective WSI label. With the rapid evolution of representation learning, numerous new feature extractor models, often termed foundational models, have emerged. Traditional evaluation methods, however, rely on fixed aggregation model hyperparameters, a framework we identify as potentially biasing the results. Our study uncovers a co-dependence between feature extractor models and aggregation model hyperparameters, indicating that performance comparability can be skewed based on the chosen hyperparameters. By accounting for this co-dependency, we find that the performance of many current feature extractor models is notably similar. We support this insight by evaluating seven feature extractor models across three different datasets with 162 different aggregation model configurations. This comprehensive approach provides a more nuanced understanding of the relationship between feature extractors and aggregation models, leading to a fairer and more accurate assessment of feature extractor models in digital pathology.
Unsupervised Domain Adaptation with Contrastive Learning for OCT Segmentation
Mar 07, 2022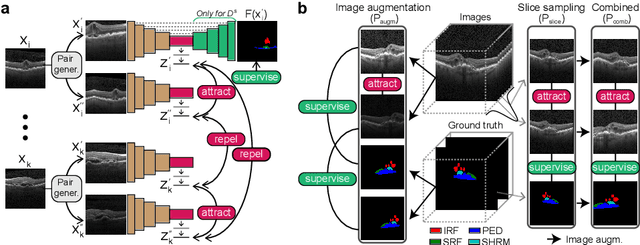
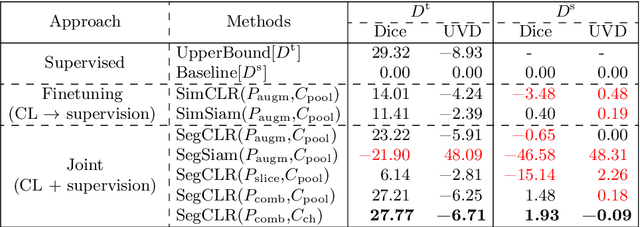
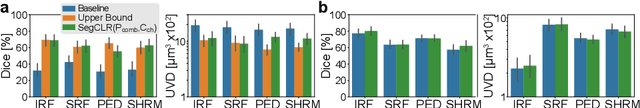
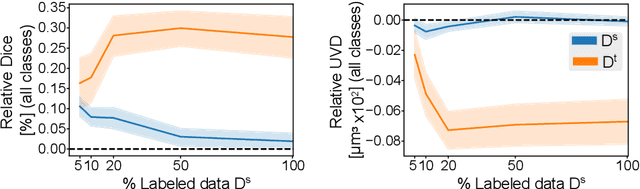
Abstract:Accurate segmentation of retinal fluids in 3D Optical Coherence Tomography images is key for diagnosis and personalized treatment of eye diseases. While deep learning has been successful at this task, trained supervised models often fail for images that do not resemble labeled examples, e.g. for images acquired using different devices. We hereby propose a novel semi-supervised learning framework for segmentation of volumetric images from new unlabeled domains. We jointly use supervised and contrastive learning, also introducing a contrastive pairing scheme that leverages similarity between nearby slices in 3D. In addition, we propose channel-wise aggregation as an alternative to conventional spatial-pooling aggregation for contrastive feature map projection. We evaluate our methods for domain adaptation from a (labeled) source domain to an (unlabeled) target domain, each containing images acquired with different acquisition devices. In the target domain, our method achieves a Dice coefficient 13.8% higher than SimCLR (a state-of-the-art contrastive framework), and leads to results comparable to an upper bound with supervised training in that domain. In the source domain, our model also improves the results by 5.4% Dice, by successfully leveraging information from many unlabeled images.
Probabilistic Spatial Analysis in Quantitative Microscopy with Uncertainty-Aware Cell Detection using Deep Bayesian Regression of Density Maps
Feb 23, 2021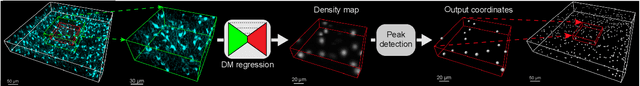

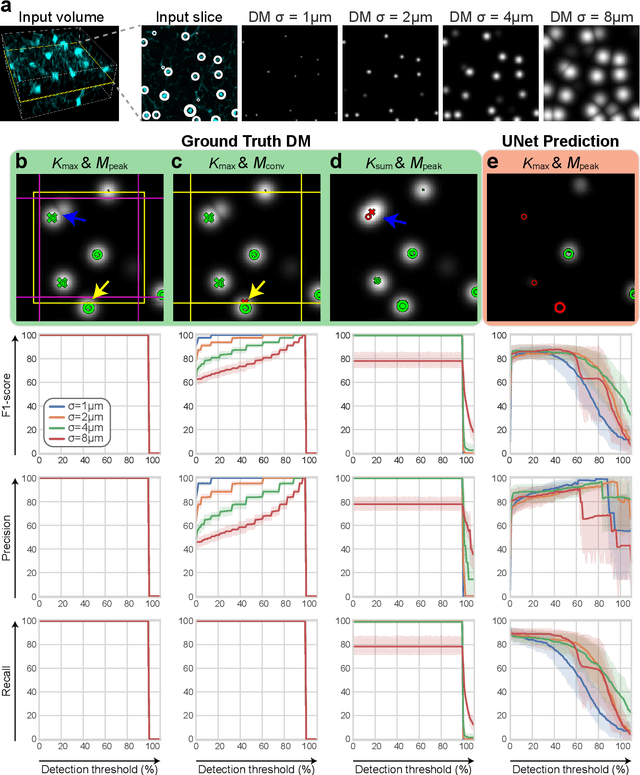
Abstract:3D microscopy is key in the investigation of diverse biological systems, and the ever increasing availability of large datasets demands automatic cell identification methods that not only are accurate, but also can imply the uncertainty in their predictions to inform about potential errors and hence confidence in conclusions using them. While conventional deep learning methods often yield deterministic results, advances in deep Bayesian learning allow for accurate predictions with a probabilistic interpretation in numerous image classification and segmentation tasks. It is however nontrivial to extend such Bayesian methods to cell detection, which requires specialized learning frameworks. In particular, regression of density maps is a popular successful approach for extracting cell coordinates from local peaks in a postprocessing step, which hinders any meaningful probabilistic output. We herein propose a deep learning-based cell detection framework that can operate on large microscopy images and outputs desired probabilistic predictions by (i) integrating Bayesian techniques for the regression of uncertainty-aware density maps, where peak detection can be applied to generate cell proposals, and (ii) learning a mapping from the numerous proposals to a probabilistic space that is calibrated, i.e. accurately represents the chances of a successful prediction. Utilizing such calibrated predictions, we propose a probabilistic spatial analysis with Monte-Carlo sampling. We demonstrate this in revising an existing description of the distribution of a mesenchymal stromal cell type within the bone marrow, where our proposed methods allow us to reveal spatial patterns that are otherwise undetectable. Introducing such probabilistic analysis in quantitative microscopy pipelines will allow for reporting confidence intervals for testing biological hypotheses of spatial distributions.
Utilizing Uncertainty Estimation in Deep Learning Segmentation of Fluorescence Microscopy Images with Missing Markers
Jan 27, 2021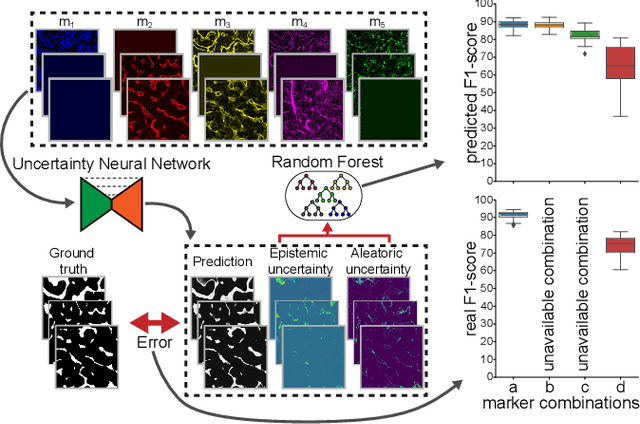
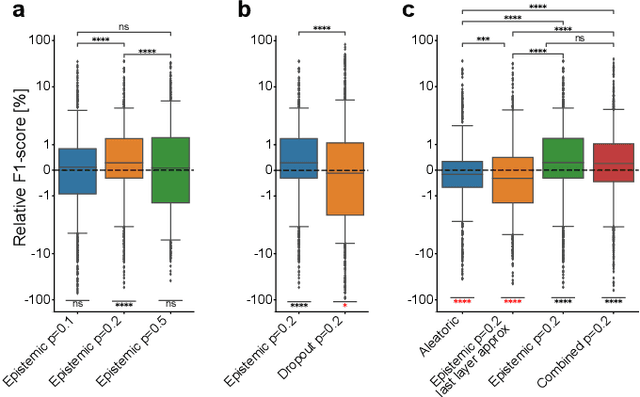
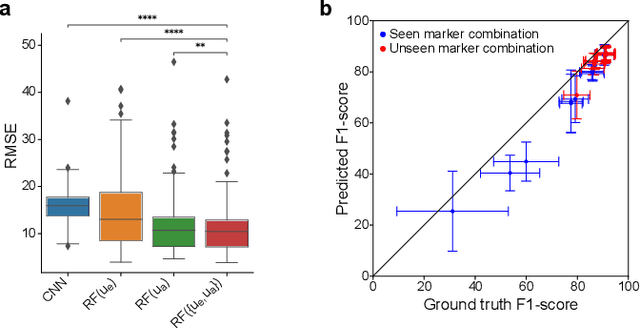
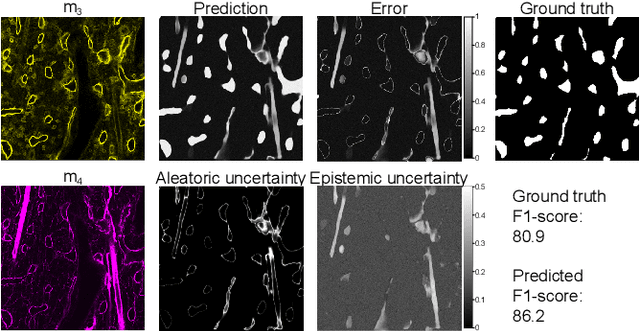
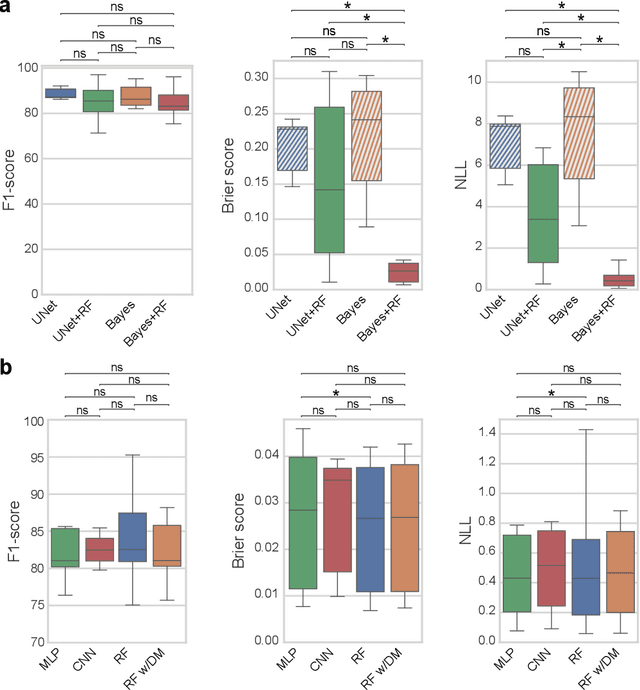
Abstract:Fluorescence microscopy images contain several channels, each indicating a marker staining the sample. Since many different marker combinations are utilized in practice, it has been challenging to apply deep learning based segmentation models, which expect a predefined channel combination for all training samples as well as at inference for future application. Recent work circumvents this problem using a modality attention approach to be effective across any possible marker combination. However, for combinations that do not exist in a labeled training dataset, one cannot have any estimation of potential segmentation quality if that combination is encountered during inference. Without this, not only one lacks quality assurance but one also does not know where to put any additional imaging and labeling effort. We herein propose a method to estimate segmentation quality on unlabeled images by (i) estimating both aleatoric and epistemic uncertainties of convolutional neural networks for image segmentation, and (ii) training a Random Forest model for the interpretation of uncertainty features via regression to their corresponding segmentation metrics. Additionally, we demonstrate that including these uncertainty measures during training can provide an improvement on segmentation performance.
Modality Attention and Sampling Enables Deep Learning with Heterogeneous Marker Combinations in Fluorescence Microscopy
Aug 27, 2020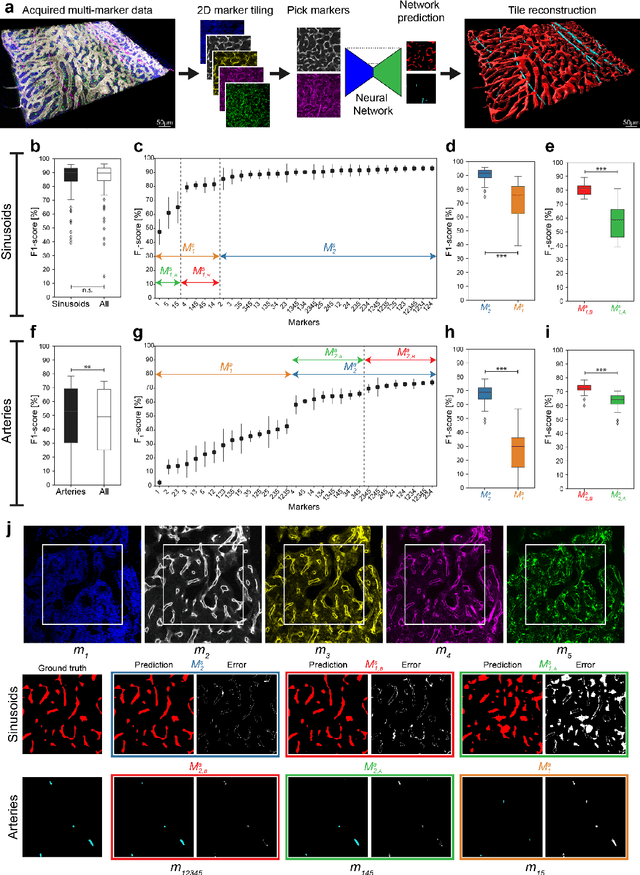

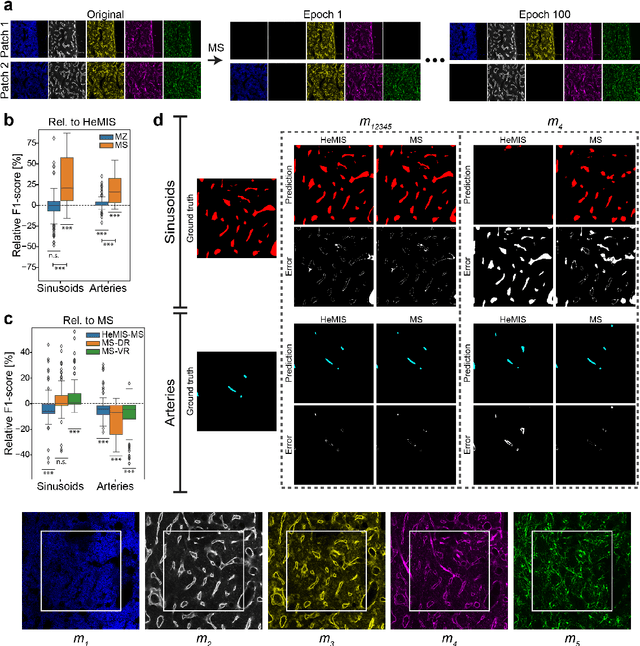
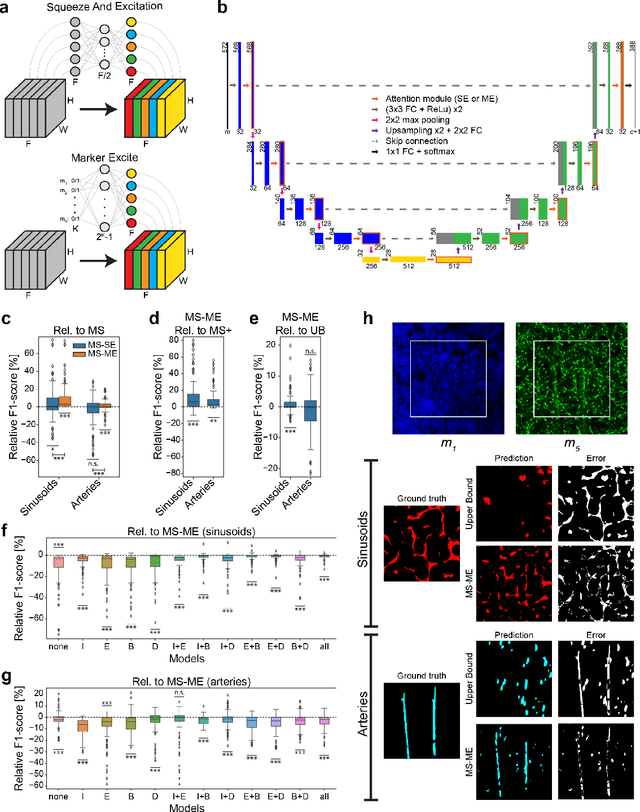
Abstract:Fluorescence microscopy allows for a detailed inspection of cells, cellular networks, and anatomical landmarks by staining with a variety of carefully-selected markers visualized as color channels. Quantitative characterization of structures in acquired images often relies on automatic image analysis methods. Despite the success of deep learning methods in other vision applications, their potential for fluorescence image analysis remains underexploited. One reason lies in the considerable workload required to train accurate models, which are normally specific for a given combination of markers, and therefore applicable to a very restricted number of experimental settings. We herein propose Marker Sampling and Excite, a neural network approach with a modality sampling strategy and a novel attention module that together enable ($i$) flexible training with heterogeneous datasets with combinations of markers and ($ii$) successful utility of learned models on arbitrary subsets of markers prospectively. We show that our single neural network solution performs comparably to an upper bound scenario where an ensemble of many networks is na\"ively trained for each possible marker combination separately. In addition, we demonstrate the feasibility of our framework in high-throughput biological analysis by revising a recent quantitative characterization of bone marrow vasculature in 3D confocal microscopy datasets. Not only can our work substantially ameliorate the use of deep learning in fluorescence microscopy analysis, but it can also be utilized in other fields with incomplete data acquisitions and missing modalities.
Siamese Networks with Location Prior for Landmark Tracking in Liver Ultrasound Sequences
Jan 23, 2019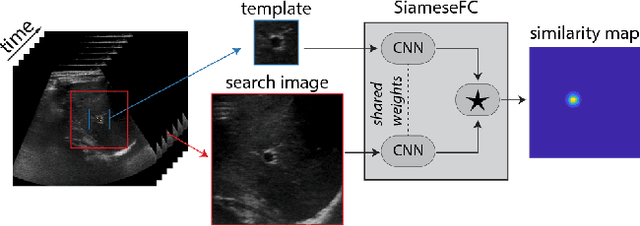
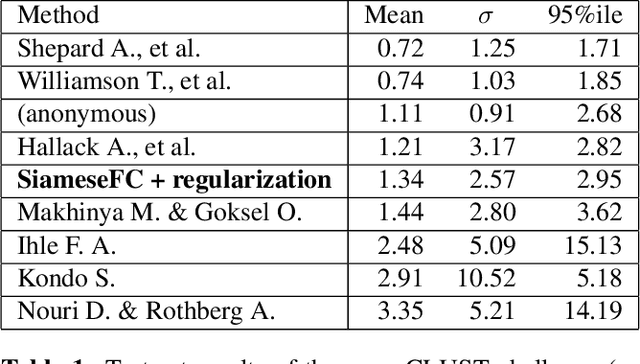
Abstract:Image-guided radiation therapy can benefit from accurate motion tracking by ultrasound imaging, in order to minimize treatment margins and radiate moving anatomical targets, e.g., due to breathing. One way to formulate this tracking problem is the automatic localization of given tracked anatomical landmarks throughout a temporal ultrasound sequence. For this, we herein propose a fully-convolutional Siamese network that learns the similarity between pairs of image regions containing the same landmark. Accordingly, it learns to localize and thus track arbitrary image features, not only predefined anatomical structures. We employ a temporal consistency model as a location prior, which we combine with the network-predicted location probability map to track a target iteratively in ultrasound sequences. We applied this method on the dataset of the Challenge on Liver Ultrasound Tracking (CLUST) with competitive results, where our work is the first to effectively apply CNNs on this tracking problem, thanks to our temporal regularization.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge