Ece Ozkan
From Slices to Structures: Unsupervised 3D Reconstruction of Female Pelvic Anatomy from Freehand Transvaginal Ultrasound
Aug 20, 2025Abstract:Volumetric ultrasound has the potential to significantly improve diagnostic accuracy and clinical decision-making, yet its widespread adoption remains limited by dependence on specialized hardware and restrictive acquisition protocols. In this work, we present a novel unsupervised framework for reconstructing 3D anatomical structures from freehand 2D transvaginal ultrasound (TVS) sweeps, without requiring external tracking or learned pose estimators. Our method adapts the principles of Gaussian Splatting to the domain of ultrasound, introducing a slice-aware, differentiable rasterizer tailored to the unique physics and geometry of ultrasound imaging. We model anatomy as a collection of anisotropic 3D Gaussians and optimize their parameters directly from image-level supervision, leveraging sensorless probe motion estimation and domain-specific geometric priors. The result is a compact, flexible, and memory-efficient volumetric representation that captures anatomical detail with high spatial fidelity. This work demonstrates that accurate 3D reconstruction from 2D ultrasound images can be achieved through purely computational means, offering a scalable alternative to conventional 3D systems and enabling new opportunities for AI-assisted analysis and diagnosis.
Multi-domain improves out-of-distribution and data-limited scenarios for medical image analysis
Oct 10, 2023Abstract:Current machine learning methods for medical image analysis primarily focus on developing models tailored for their specific tasks, utilizing data within their target domain. These specialized models tend to be data-hungry and often exhibit limitations in generalizing to out-of-distribution samples. Recently, foundation models have been proposed, which combine data from various domains and demonstrate excellent generalization capabilities. Building upon this, this work introduces the incorporation of diverse medical image domains, including different imaging modalities like X-ray, MRI, CT, and ultrasound images, as well as various viewpoints such as axial, coronal, and sagittal views. We refer to this approach as multi-domain model and compare its performance to that of specialized models. Our findings underscore the superior generalization capabilities of multi-domain models, particularly in scenarios characterized by limited data availability and out-of-distribution, frequently encountered in healthcare applications. The integration of diverse data allows multi-domain models to utilize shared information across domains, enhancing the overall outcomes significantly. To illustrate, for organ recognition, multi-domain model can enhance accuracy by up to 10% compared to conventional specialized models.
M-mode Based Prediction of Ejection Fraction using Echocardiograms
Sep 07, 2023



Abstract:Early detection of cardiac dysfunction through routine screening is vital for diagnosing cardiovascular diseases. An important metric of cardiac function is the left ventricular ejection fraction (EF), where lower EF is associated with cardiomyopathy. Echocardiography is a popular diagnostic tool in cardiology, with ultrasound being a low-cost, real-time, and non-ionizing technology. However, human assessment of echocardiograms for calculating EF is time-consuming and expertise-demanding, raising the need for an automated approach. In this work, we propose using the M(otion)-mode of echocardiograms for estimating the EF and classifying cardiomyopathy. We generate multiple artificial M-mode images from a single echocardiogram and combine them using off-the-shelf model architectures. Additionally, we extend contrastive learning (CL) to cardiac imaging to learn meaningful representations from exploiting structures in unlabeled data allowing the model to achieve high accuracy, even with limited annotations. Our experiments show that the supervised setting converges with only ten modes and is comparable to the baseline method while bypassing its cumbersome training process and being computationally much more efficient. Furthermore, CL using M-mode images is helpful for limited data scenarios, such as having labels for only 200 patients, which is common in medical applications.
Interpretable and Intervenable Ultrasonography-based Machine Learning Models for Pediatric Appendicitis
Feb 28, 2023Abstract:Appendicitis is among the most frequent reasons for pediatric abdominal surgeries. With recent advances in machine learning, data-driven decision support could help clinicians diagnose and manage patients while reducing the number of non-critical surgeries. Previous decision support systems for appendicitis focused on clinical, laboratory, scoring and computed tomography data, mainly ignoring abdominal ultrasound, a noninvasive and readily available diagnostic modality. To this end, we developed and validated interpretable machine learning models for predicting the diagnosis, management and severity of suspected appendicitis using ultrasound images. Our models were trained on a dataset comprising 579 pediatric patients with 1709 ultrasound images accompanied by clinical and laboratory data. Our methodological contribution is the generalization of concept bottleneck models to prediction problems with multiple views and incomplete concept sets. Notably, such models lend themselves to interpretation and interaction via high-level concepts understandable to clinicians without sacrificing performance or requiring time-consuming image annotation when deployed.
Introduction to Machine Learning for Physicians: A Survival Guide for Data Deluge
Dec 23, 2022
Abstract:Many modern research fields increasingly rely on collecting and analysing massive, often unstructured, and unwieldy datasets. Consequently, there is growing interest in machine learning and artificial intelligence applications that can harness this `data deluge'. This broad nontechnical overview provides a gentle introduction to machine learning with a specific focus on medical and biological applications. We explain the common types of machine learning algorithms and typical tasks that can be solved, illustrating the basics with concrete examples from healthcare. Lastly, we provide an outlook on open challenges, limitations, and potential impacts of machine-learning-powered medicine.
Debiasing Deep Chest X-Ray Classifiers using Intra- and Post-processing Methods
Jul 26, 2022



Abstract:Deep neural networks for image-based screening and computer-aided diagnosis have achieved expert-level performance on various medical imaging modalities, including chest radiographs. Recently, several works have indicated that these state-of-the-art classifiers can be biased with respect to sensitive patient attributes, such as race or gender, leading to growing concerns about demographic disparities and discrimination resulting from algorithmic and model-based decision-making in healthcare. Fair machine learning has focused on mitigating such biases against disadvantaged or marginalised groups, mainly concentrating on tabular data or natural images. This work presents two novel intra-processing techniques based on fine-tuning and pruning an already-trained neural network. These methods are simple yet effective and can be readily applied post hoc in a setting where the protected attribute is unknown during the model development and test time. In addition, we compare several intra- and post-processing approaches applied to debiasing deep chest X-ray classifiers. To the best of our knowledge, this is one of the first efforts studying debiasing methods on chest radiographs. Our results suggest that the considered approaches successfully mitigate biases in fully connected and convolutional neural networks offering stable performance under various settings. The discussed methods can help achieve group fairness of deep medical image classifiers when deploying them in domains with different fairness considerations and constraints.
Interpretable Prediction of Pulmonary Hypertension in Newborns using Echocardiograms
Mar 24, 2022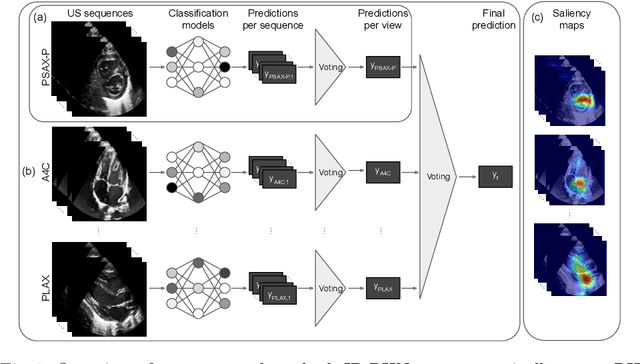
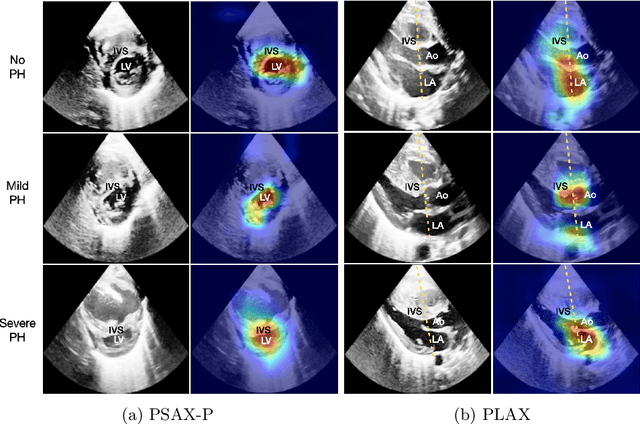
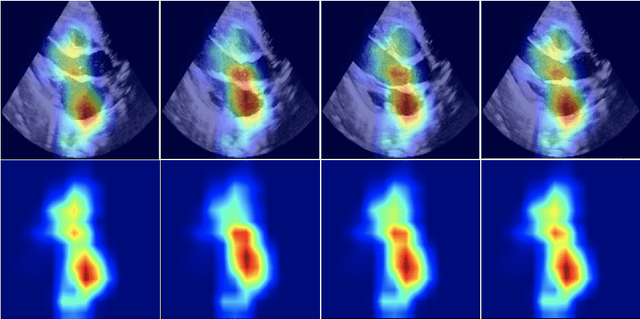
Abstract:Pulmonary hypertension (PH) in newborns and infants is a complex condition associated with several pulmonary, cardiac, and systemic diseases contributing to morbidity and mortality. Therefore, accurate and early detection of PH is crucial for successful management. Using echocardiography, the primary diagnostic tool in pediatrics, human assessment is both time-consuming and expertise-demanding, raising the need for an automated approach. In this work, we present an interpretable multi-view video-based deep learning approach to predict PH for a cohort of 194 newborns using echocardiograms. We use spatio-temporal convolutional architectures for the prediction of PH from each view, and aggregate the predictions of the different views using majority voting. To the best of our knowledge, this is the first work for an automated assessment of PH in newborns using echocardiograms. Our results show a mean F1-score of 0.84 for severity prediction and 0.92 for binary detection using 10-fold cross-validation. We complement our predictions with saliency maps and show that the learned model focuses on clinically relevant cardiac structures, motivating its usage in clinical practice.
Siamese Networks with Location Prior for Landmark Tracking in Liver Ultrasound Sequences
Jan 23, 2019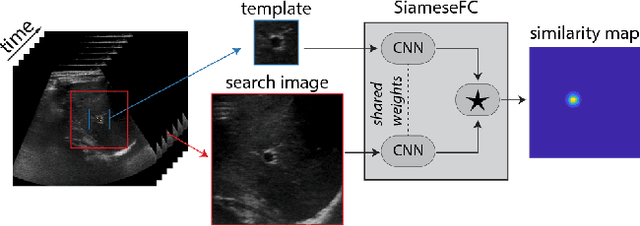
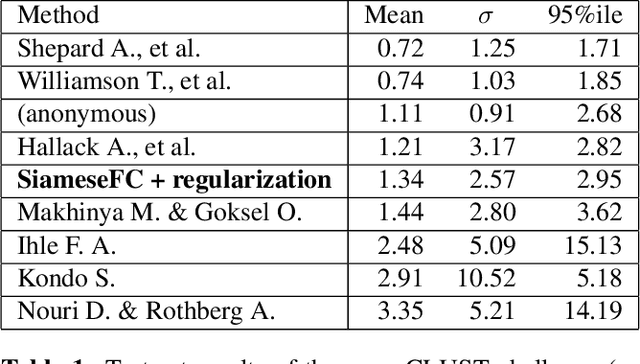
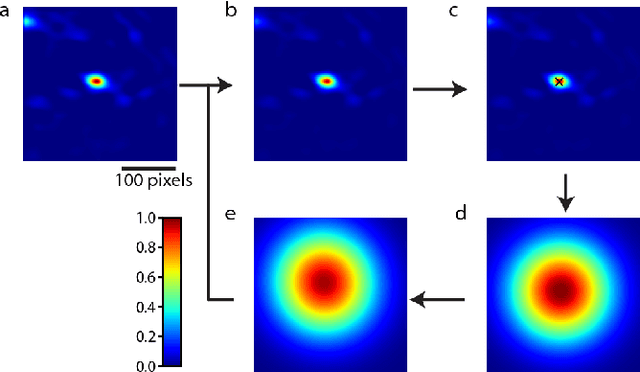
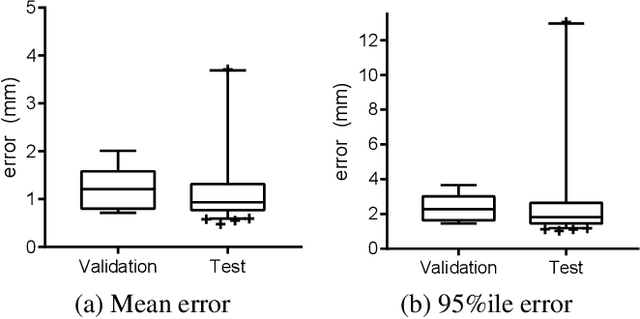
Abstract:Image-guided radiation therapy can benefit from accurate motion tracking by ultrasound imaging, in order to minimize treatment margins and radiate moving anatomical targets, e.g., due to breathing. One way to formulate this tracking problem is the automatic localization of given tracked anatomical landmarks throughout a temporal ultrasound sequence. For this, we herein propose a fully-convolutional Siamese network that learns the similarity between pairs of image regions containing the same landmark. Accordingly, it learns to localize and thus track arbitrary image features, not only predefined anatomical structures. We employ a temporal consistency model as a location prior, which we combine with the network-predicted location probability map to track a target iteratively in ultrasound sequences. We applied this method on the dataset of the Challenge on Liver Ultrasound Tracking (CLUST) with competitive results, where our work is the first to effectively apply CNNs on this tracking problem, thanks to our temporal regularization.
Herding Generalizes Diverse M -Best Solutions
Jan 30, 2017



Abstract:We show that the algorithm to extract diverse M -solutions from a Conditional Random Field (called divMbest [1]) takes exactly the form of a Herding procedure [2], i.e. a deterministic dynamical system that produces a sequence of hypotheses that respect a set of observed moment constraints. This generalization enables us to invoke properties of Herding that show that divMbest enforces implausible constraints which may yield wrong assumptions for some problem settings. Our experiments in semantic segmentation demonstrate that seeing divMbest as an instance of Herding leads to better alternatives for the implausible constraints of divMbest.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge