Laura Manduchi
Leveraging Cardiovascular Simulations for In-Vivo Prediction of Cardiac Biomarkers
Dec 23, 2024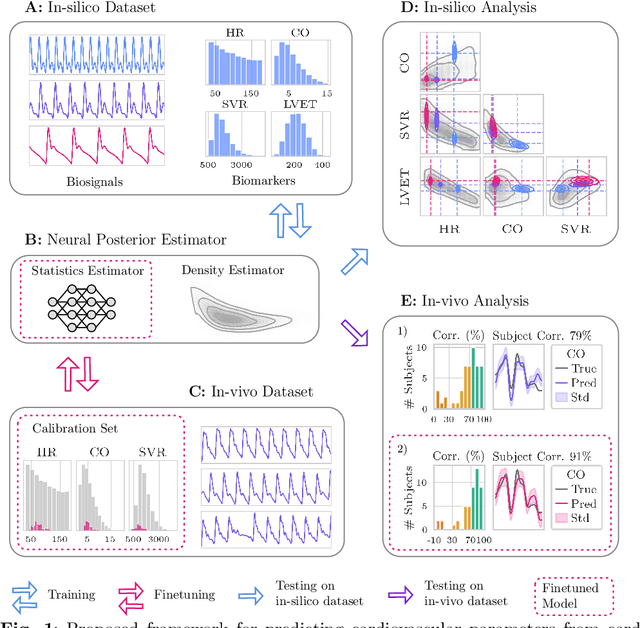
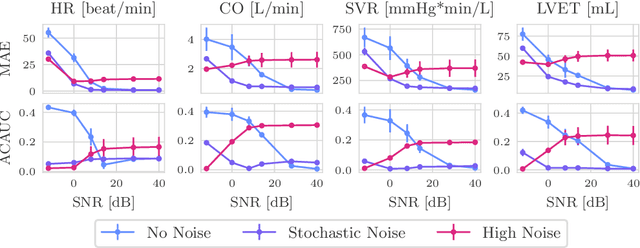
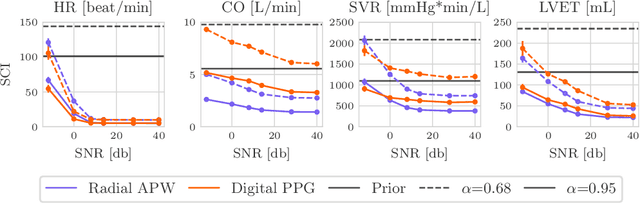
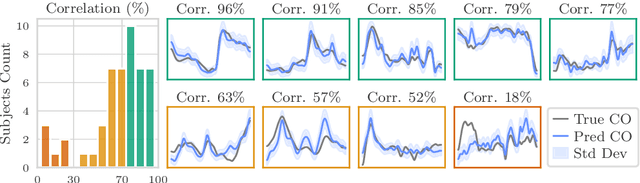
Abstract:Whole-body hemodynamics simulators, which model blood flow and pressure waveforms as functions of physiological parameters, are now essential tools for studying cardiovascular systems. However, solving the corresponding inverse problem of mapping observations (e.g., arterial pressure waveforms at specific locations in the arterial network) back to plausible physiological parameters remains challenging. Leveraging recent advances in simulation-based inference, we cast this problem as statistical inference by training an amortized neural posterior estimator on a newly built large dataset of cardiac simulations that we publicly release. To better align simulated data with real-world measurements, we incorporate stochastic elements modeling exogenous effects. The proposed framework can further integrate in-vivo data sources to refine its predictive capabilities on real-world data. In silico, we demonstrate that the proposed framework enables finely quantifying uncertainty associated with individual measurements, allowing trustworthy prediction of four biomarkers of clinical interest--namely Heart Rate, Cardiac Output, Systemic Vascular Resistance, and Left Ventricular Ejection Time--from arterial pressure waveforms and photoplethysmograms. Furthermore, we validate the framework in vivo, where our method accurately captures temporal trends in CO and SVR monitoring on the VitalDB dataset. Finally, the predictive error made by the model monotonically increases with the predicted uncertainty, thereby directly supporting the automatic rejection of unusable measurements.
Hierarchical Clustering for Conditional Diffusion in Image Generation
Oct 22, 2024



Abstract:Finding clusters of data points with similar characteristics and generating new cluster-specific samples can significantly enhance our understanding of complex data distributions. While clustering has been widely explored using Variational Autoencoders, these models often lack generation quality in real-world datasets. This paper addresses this gap by introducing TreeDiffusion, a deep generative model that conditions Diffusion Models on hierarchical clusters to obtain high-quality, cluster-specific generations. The proposed pipeline consists of two steps: a VAE-based clustering model that learns the hierarchical structure of the data, and a conditional diffusion model that generates realistic images for each cluster. We propose this two-stage process to ensure that the generated samples remain representative of their respective clusters and enhance image fidelity to the level of diffusion models. A key strength of our method is its ability to create images for each cluster, providing better visualization of the learned representations by the clustering model, as demonstrated through qualitative results. This method effectively addresses the generative limitations of VAE-based approaches while preserving their clustering performance. Empirically, we demonstrate that conditioning diffusion models on hierarchical clusters significantly enhances generative performance, thereby advancing the state of generative clustering models.
Structured Generations: Using Hierarchical Clusters to guide Diffusion Models
Jul 08, 2024Abstract:This paper introduces Diffuse-TreeVAE, a deep generative model that integrates hierarchical clustering into the framework of Denoising Diffusion Probabilistic Models (DDPMs). The proposed approach generates new images by sampling from a root embedding of a learned latent tree VAE-based structure, it then propagates through hierarchical paths, and utilizes a second-stage DDPM to refine and generate distinct, high-quality images for each data cluster. The result is a model that not only improves image clarity but also ensures that the generated samples are representative of their respective clusters, addressing the limitations of previous VAE-based methods and advancing the state of clustering-based generative modeling.
scTree: Discovering Cellular Hierarchies in the Presence of Batch Effects in scRNA-seq Data
Jun 27, 2024



Abstract:We propose a novel method, scTree, for single-cell Tree Variational Autoencoders, extending a hierarchical clustering approach to single-cell RNA sequencing data. scTree corrects for batch effects while simultaneously learning a tree-structured data representation. This VAE-based method allows for a more in-depth understanding of complex cellular landscapes independently of the biasing effects of batches. We show empirically on seven datasets that scTree discovers the underlying clusters of the data and the hierarchical relations between them, as well as outperforms established baseline methods across these datasets. Additionally, we analyze the learned hierarchy to understand its biological relevance, thus underpinning the importance of integrating batch correction directly into the clustering procedure.
On the Challenges and Opportunities in Generative AI
Feb 28, 2024Abstract:The field of deep generative modeling has grown rapidly and consistently over the years. With the availability of massive amounts of training data coupled with advances in scalable unsupervised learning paradigms, recent large-scale generative models show tremendous promise in synthesizing high-resolution images and text, as well as structured data such as videos and molecules. However, we argue that current large-scale generative AI models do not sufficiently address several fundamental issues that hinder their widespread adoption across domains. In this work, we aim to identify key unresolved challenges in modern generative AI paradigms that should be tackled to further enhance their capabilities, versatility, and reliability. By identifying these challenges, we aim to provide researchers with valuable insights for exploring fruitful research directions, thereby fostering the development of more robust and accessible generative AI solutions.
Tree Variational Autoencoders
Jul 01, 2023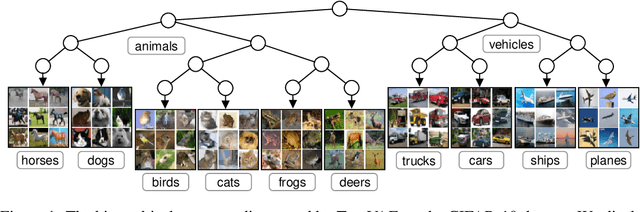
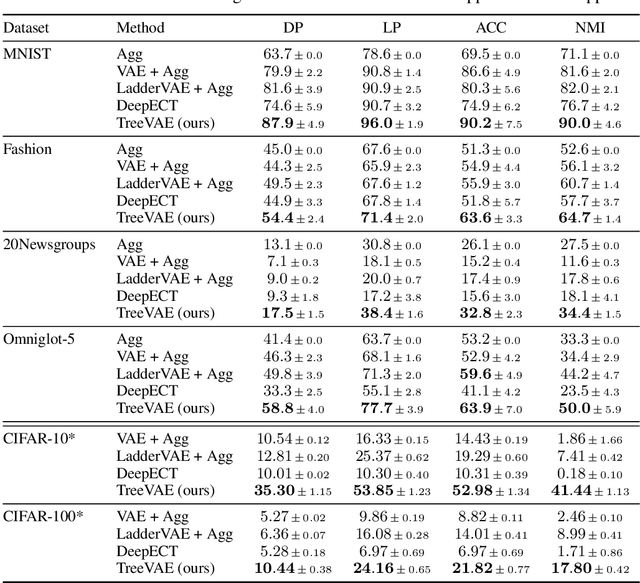
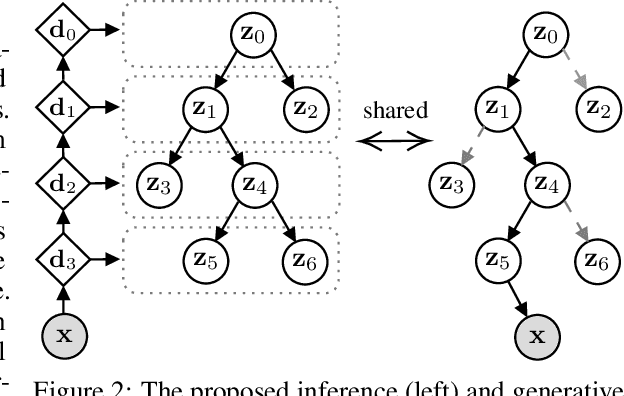
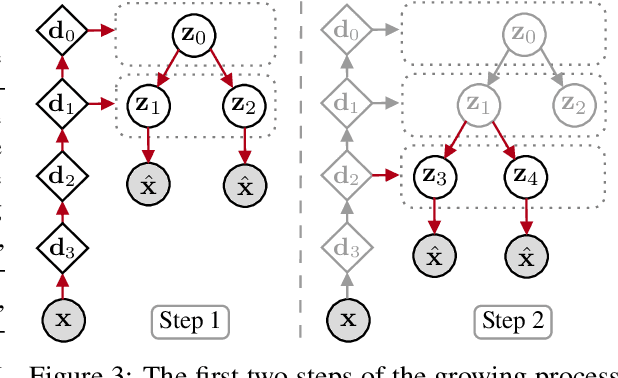
Abstract:We propose a new generative hierarchical clustering model that learns a flexible tree-based posterior distribution over latent variables. The proposed Tree Variational Autoencoder (TreeVAE) hierarchically divides samples according to their intrinsic characteristics, shedding light on hidden structure in the data. It adapts its architecture to discover the optimal tree for encoding dependencies between latent variables. The proposed tree-based generative architecture permits lightweight conditional inference and improves generative performance by utilizing specialized leaf decoders. We show that TreeVAE uncovers underlying clusters in the data and finds meaningful hierarchical relations between the different groups on a variety of datasets, including real-world imaging data. We present empirically that TreeVAE provides a more competitive log-likelihood lower bound than the sequential counterparts. Finally, due to its generative nature, TreeVAE is able to generate new samples from the discovered clusters via conditional sampling.
Signal Is Harder To Learn Than Bias: Debiasing with Focal Loss
May 31, 2023Abstract:Spurious correlations are everywhere. While humans often do not perceive them, neural networks are notorious for learning unwanted associations, also known as biases, instead of the underlying decision rule. As a result, practitioners are often unaware of the biased decision-making of their classifiers. Such a biased model based on spurious correlations might not generalize to unobserved data, leading to unintended, adverse consequences. We propose Signal is Harder (SiH), a variational-autoencoder-based method that simultaneously trains a biased and unbiased classifier using a novel, disentangling reweighting scheme inspired by the focal loss. Using the unbiased classifier, SiH matches or improves upon the performance of state-of-the-art debiasing methods. To improve the interpretability of our technique, we propose a perturbation scheme in the latent space for visualizing the bias that helps practitioners become aware of the sources of spurious correlations.
Interpretable Anomaly Detection in Echocardiograms with Dynamic Variational Trajectory Models
Jun 30, 2022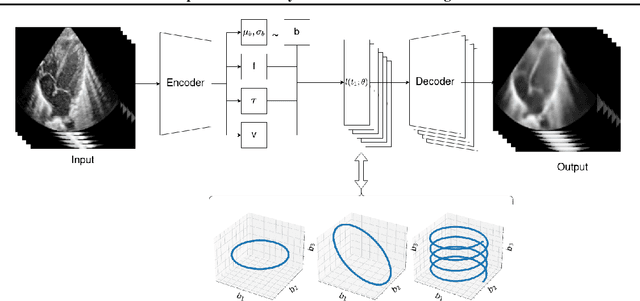
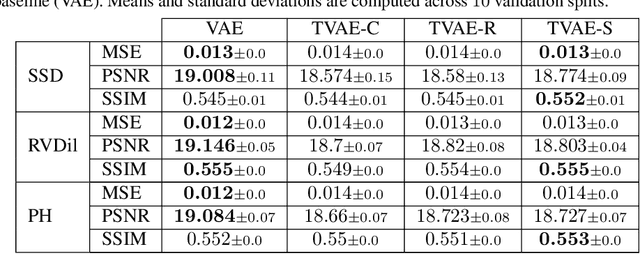
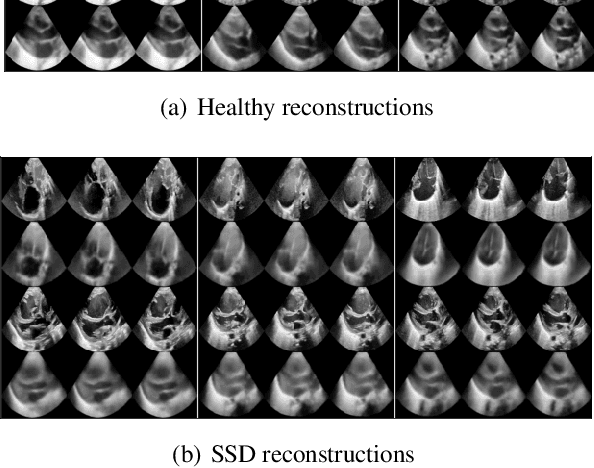
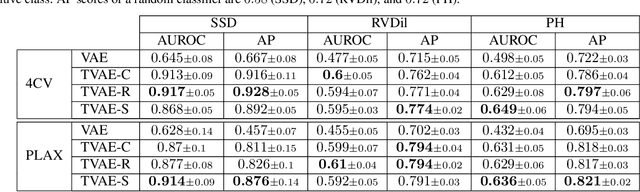
Abstract:We propose a novel anomaly detection method for echocardiogram videos. The introduced method takes advantage of the periodic nature of the heart cycle to learn different variants of a variational latent trajectory model (TVAE). The models are trained on the healthy samples of an in-house dataset of infant echocardiogram videos consisting of multiple chamber views to learn a normative prior of the healthy population. During inference, maximum a posteriori (MAP) based anomaly detection is performed to detect out-of-distribution samples in our dataset. The proposed method reliably identifies severe congenital heart defects, such as Ebstein's Anomaly or Shonecomplex. Moreover, it achieves superior performance over MAP-based anomaly detection with standard variational autoencoders on the task of detecting pulmonary hypertension and right ventricular dilation. Finally, we demonstrate that the proposed method provides interpretable explanations of its output through heatmaps which highlight the regions corresponding to anomalous heart structures.
Interpretable Prediction of Pulmonary Hypertension in Newborns using Echocardiograms
Mar 24, 2022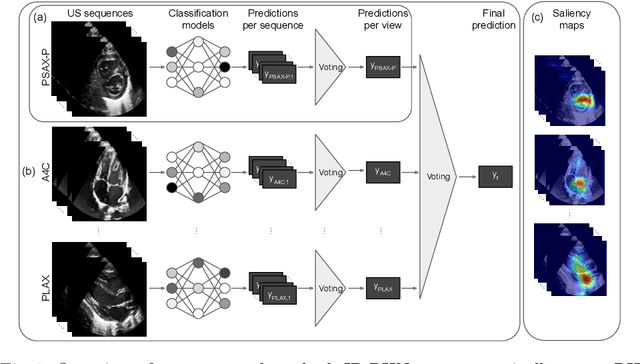
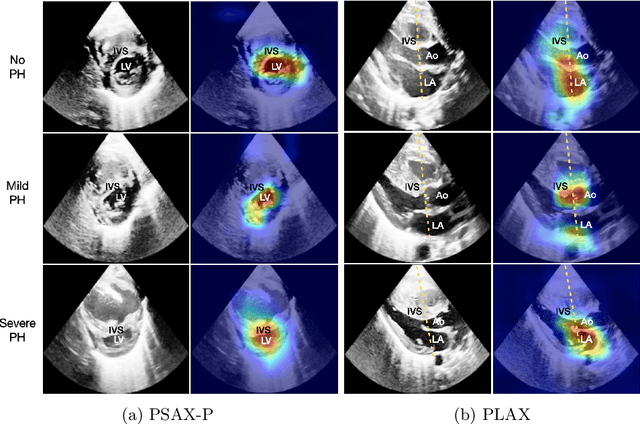
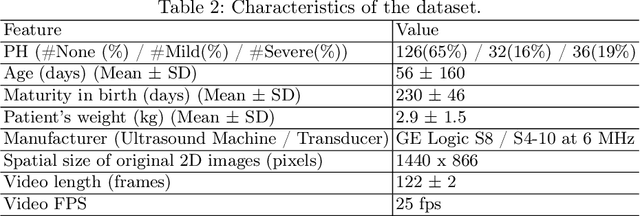
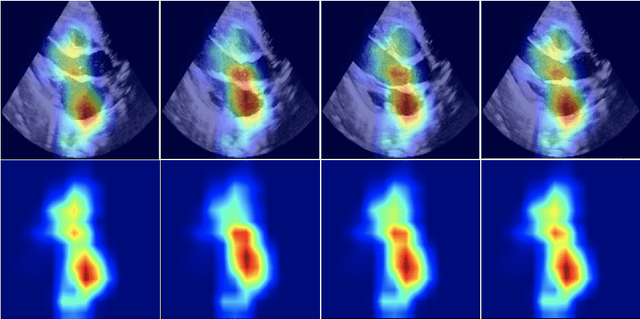
Abstract:Pulmonary hypertension (PH) in newborns and infants is a complex condition associated with several pulmonary, cardiac, and systemic diseases contributing to morbidity and mortality. Therefore, accurate and early detection of PH is crucial for successful management. Using echocardiography, the primary diagnostic tool in pediatrics, human assessment is both time-consuming and expertise-demanding, raising the need for an automated approach. In this work, we present an interpretable multi-view video-based deep learning approach to predict PH for a cohort of 194 newborns using echocardiograms. We use spatio-temporal convolutional architectures for the prediction of PH from each view, and aggregate the predictions of the different views using majority voting. To the best of our knowledge, this is the first work for an automated assessment of PH in newborns using echocardiograms. Our results show a mean F1-score of 0.84 for severity prediction and 0.92 for binary detection using 10-fold cross-validation. We complement our predictions with saliency maps and show that the learned model focuses on clinically relevant cardiac structures, motivating its usage in clinical practice.
Continuous Relaxation For The Multivariate Non-Central Hypergeometric Distribution
Mar 03, 2022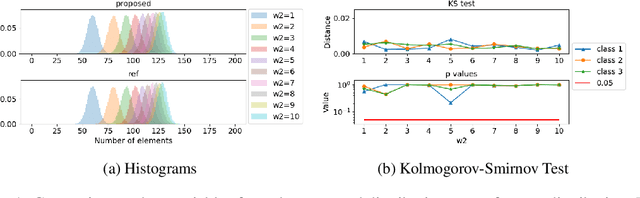
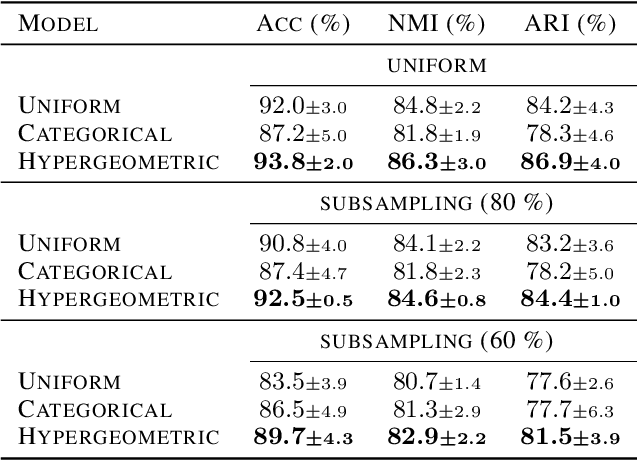
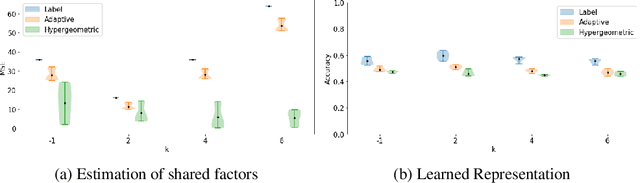
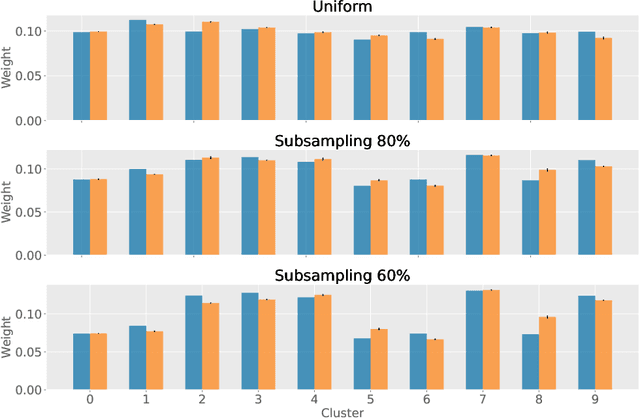
Abstract:Partitioning a set of elements into a given number of groups of a priori unknown sizes is an important task in many applications. Due to hard constraints, it is a non-differentiable problem which prohibits its direct use in modern machine learning frameworks. Hence, previous works mostly fall back on suboptimal heuristics or simplified assumptions. The multivariate hypergeometric distribution offers a probabilistic formulation of how to distribute a given number of samples across multiple groups. Unfortunately, as a discrete probability distribution, it neither is differentiable. In this work, we propose a continuous relaxation for the multivariate non-central hypergeometric distribution. We introduce an efficient and numerically stable sampling procedure. This enables reparameterized gradients for the hypergeometric distribution and its integration into automatic differentiation frameworks. We highlight the applicability and usability of the proposed formulation on two different common machine learning tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge