Jörn-Henrik Jacobsen
IvI
Inferring Optical Tissue Properties from Photoplethysmography using Hybrid Amortized Inference
Oct 02, 2025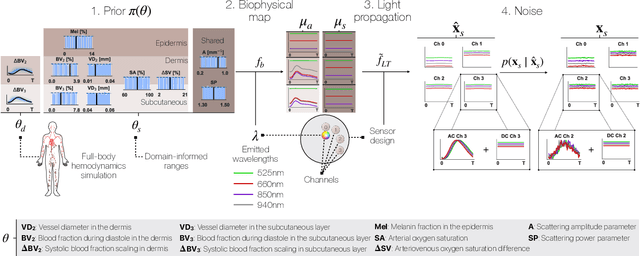
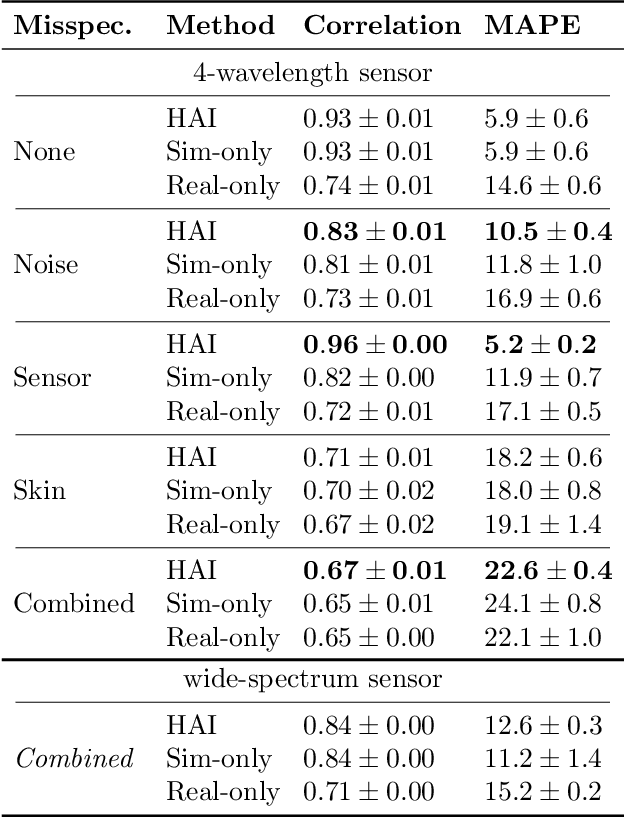
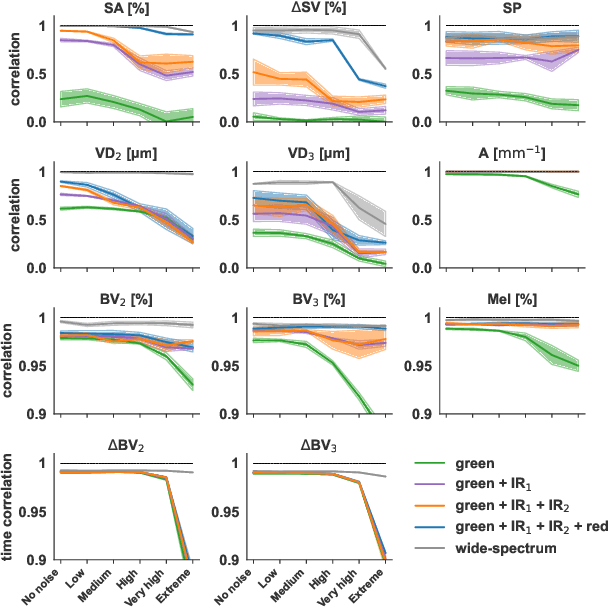

Abstract:Smart wearables enable continuous tracking of established biomarkers such as heart rate, heart rate variability, and blood oxygen saturation via photoplethysmography (PPG). Beyond these metrics, PPG waveforms contain richer physiological information, as recent deep learning (DL) studies demonstrate. However, DL models often rely on features with unclear physiological meaning, creating a tension between predictive power, clinical interpretability, and sensor design. We address this gap by introducing PPGen, a biophysical model that relates PPG signals to interpretable physiological and optical parameters. Building on PPGen, we propose hybrid amortized inference (HAI), enabling fast, robust, and scalable estimation of relevant physiological parameters from PPG signals while correcting for model misspecification. In extensive in-silico experiments, we show that HAI can accurately infer physiological parameters under diverse noise and sensor conditions. Our results illustrate a path toward PPG models that retain the fidelity needed for DL-based features while supporting clinical interpretation and informed hardware design.
Leveraging Cardiovascular Simulations for In-Vivo Prediction of Cardiac Biomarkers
Dec 23, 2024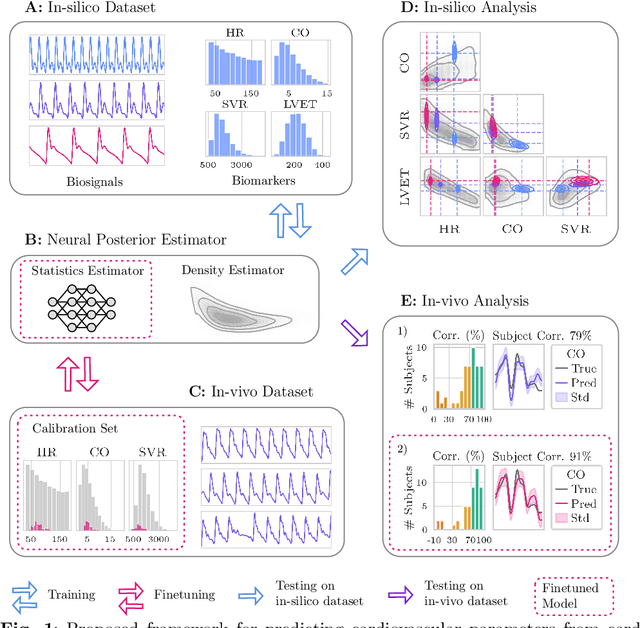
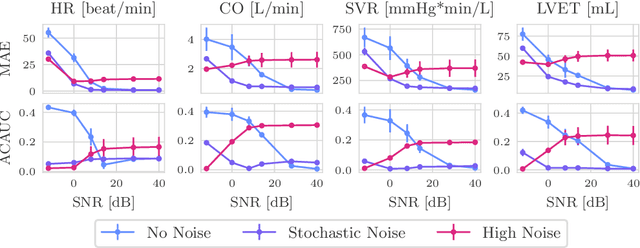
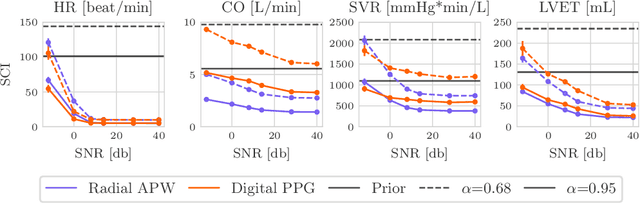
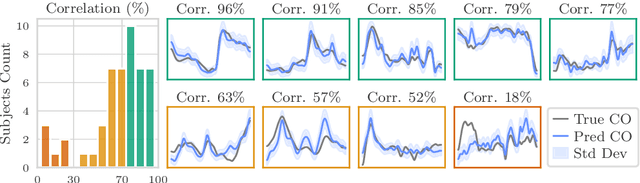
Abstract:Whole-body hemodynamics simulators, which model blood flow and pressure waveforms as functions of physiological parameters, are now essential tools for studying cardiovascular systems. However, solving the corresponding inverse problem of mapping observations (e.g., arterial pressure waveforms at specific locations in the arterial network) back to plausible physiological parameters remains challenging. Leveraging recent advances in simulation-based inference, we cast this problem as statistical inference by training an amortized neural posterior estimator on a newly built large dataset of cardiac simulations that we publicly release. To better align simulated data with real-world measurements, we incorporate stochastic elements modeling exogenous effects. The proposed framework can further integrate in-vivo data sources to refine its predictive capabilities on real-world data. In silico, we demonstrate that the proposed framework enables finely quantifying uncertainty associated with individual measurements, allowing trustworthy prediction of four biomarkers of clinical interest--namely Heart Rate, Cardiac Output, Systemic Vascular Resistance, and Left Ventricular Ejection Time--from arterial pressure waveforms and photoplethysmograms. Furthermore, we validate the framework in vivo, where our method accurately captures temporal trends in CO and SVR monitoring on the VitalDB dataset. Finally, the predictive error made by the model monotonically increases with the predicted uncertainty, thereby directly supporting the automatic rejection of unusable measurements.
Considerations for Distribution Shift Robustness of Diagnostic Models in Healthcare
Oct 25, 2024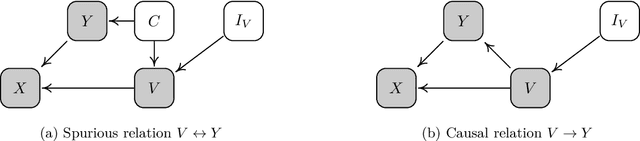
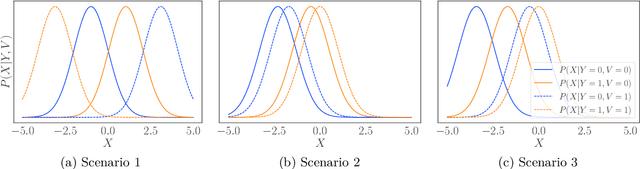
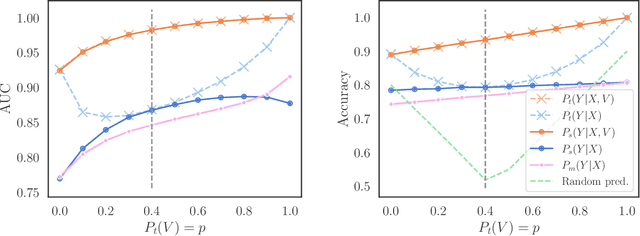
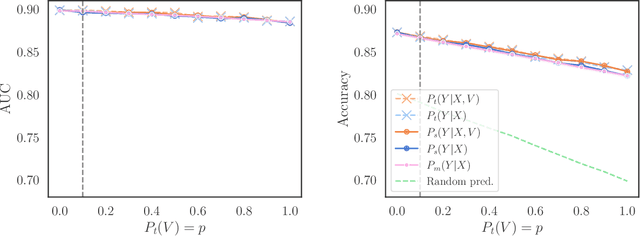
Abstract:We consider robustness to distribution shifts in the context of diagnostic models in healthcare, where the prediction target $Y$, e.g., the presence of a disease, is causally upstream of the observations $X$, e.g., a biomarker. Distribution shifts may occur, for instance, when the training data is collected in a domain with patients having particular demographic characteristics while the model is deployed on patients from a different demographic group. In the domain of applied ML for health, it is common to predict $Y$ from $X$ without considering further information about the patient. However, beyond the direct influence of the disease $Y$ on biomarker $X$, a predictive model may learn to exploit confounding dependencies (or shortcuts) between $X$ and $Y$ that are unstable under certain distribution shifts. In this work, we highlight a data generating mechanism common to healthcare settings and discuss how recent theoretical results from the causality literature can be applied to build robust predictive models. We theoretically show why ignoring covariates as well as common invariant learning approaches will in general not yield robust predictors in the studied setting, while including certain covariates into the prediction model will. In an extensive simulation study, we showcase the robustness (or lack thereof) of different predictors under various data generating processes. Lastly, we analyze the performance of the different approaches using the PTB-XL dataset, a public dataset of annotated ECG recordings.
Addressing Misspecification in Simulation-based Inference through Data-driven Calibration
May 14, 2024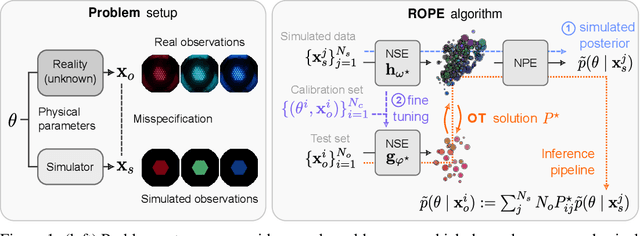
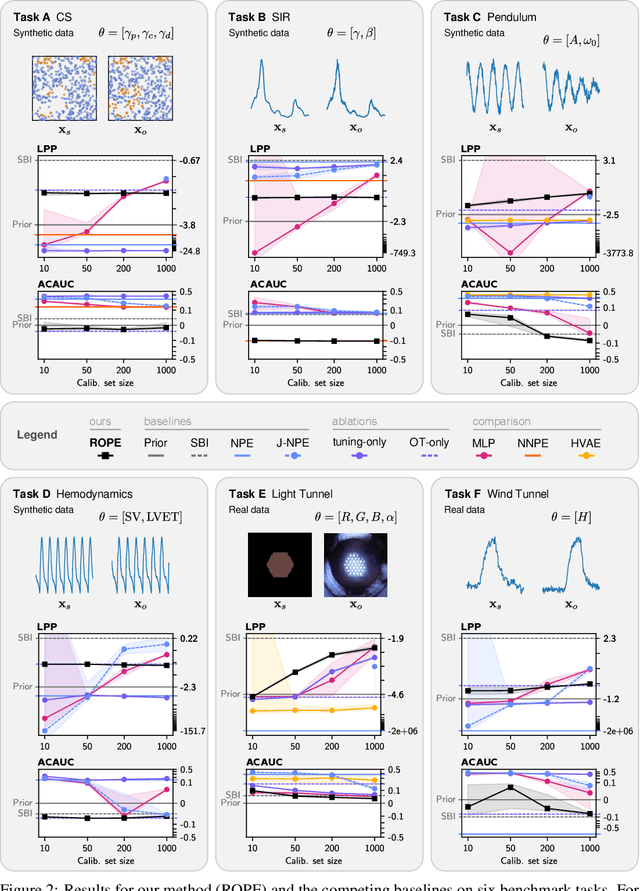
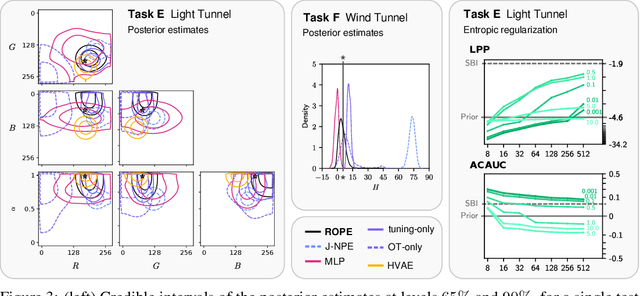
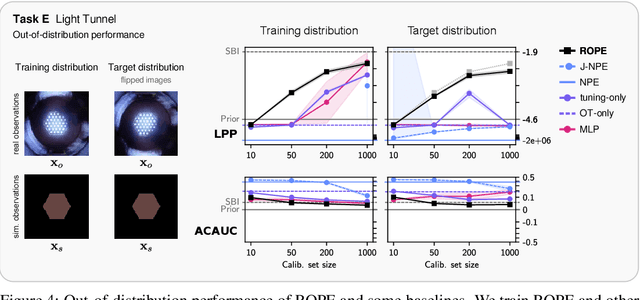
Abstract:Driven by steady progress in generative modeling, simulation-based inference (SBI) has enabled inference over stochastic simulators. However, recent work has demonstrated that model misspecification can harm SBI's reliability. This work introduces robust posterior estimation (ROPE), a framework that overcomes model misspecification with a small real-world calibration set of ground truth parameter measurements. We formalize the misspecification gap as the solution of an optimal transport problem between learned representations of real-world and simulated observations. Assuming the prior distribution over the parameters of interest is known and well-specified, our method offers a controllable balance between calibrated uncertainty and informative inference under all possible misspecifications of the simulator. Our empirical results on four synthetic tasks and two real-world problems demonstrate that ROPE outperforms baselines and consistently returns informative and calibrated credible intervals.
Simulation-based Inference for Cardiovascular Models
Jul 29, 2023Abstract:Over the past decades, hemodynamics simulators have steadily evolved and have become tools of choice for studying cardiovascular systems in-silico. While such tools are routinely used to simulate whole-body hemodynamics from physiological parameters, solving the corresponding inverse problem of mapping waveforms back to plausible physiological parameters remains both promising and challenging. Motivated by advances in simulation-based inference (SBI), we cast this inverse problem as statistical inference. In contrast to alternative approaches, SBI provides \textit{posterior distributions} for the parameters of interest, providing a \textit{multi-dimensional} representation of uncertainty for \textit{individual} measurements. We showcase this ability by performing an in-silico uncertainty analysis of five biomarkers of clinical interest comparing several measurement modalities. Beyond the corroboration of known facts, such as the feasibility of estimating heart rate, our study highlights the potential of estimating new biomarkers from standard-of-care measurements. SBI reveals practically relevant findings that cannot be captured by standard sensitivity analyses, such as the existence of sub-populations for which parameter estimation exhibits distinct uncertainty regimes. Finally, we study the gap between in-vivo and in-silico with the MIMIC-III waveform database and critically discuss how cardiovascular simulations can inform real-world data analysis.
Robust Hybrid Learning With Expert Augmentation
Feb 09, 2022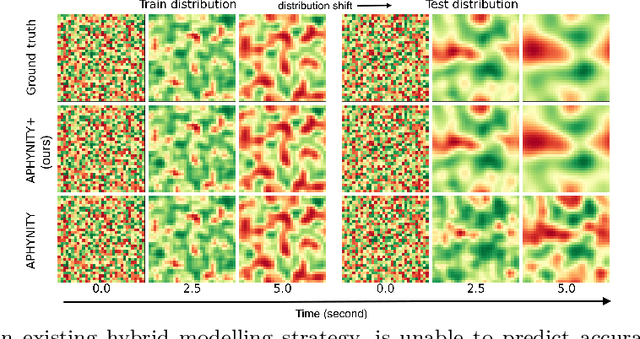
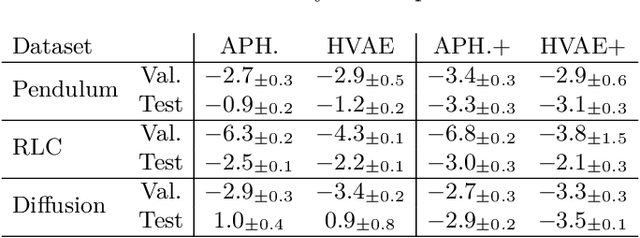
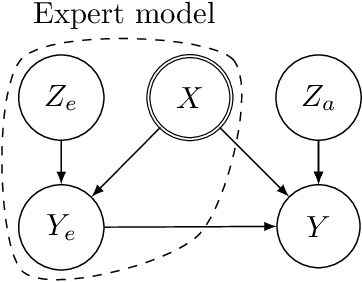
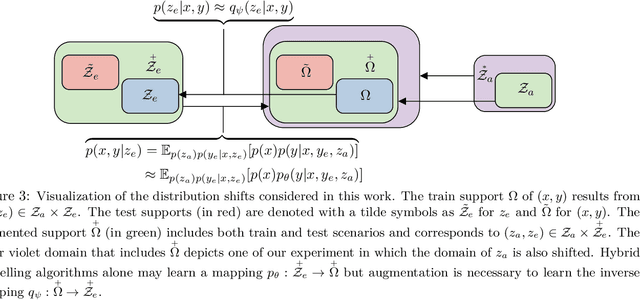
Abstract:Hybrid modelling reduces the misspecification of expert models by combining them with machine learning (ML) components learned from data. Like for many ML algorithms, hybrid model performance guarantees are limited to the training distribution. Leveraging the insight that the expert model is usually valid even outside the training domain, we overcome this limitation by introducing a hybrid data augmentation strategy termed \textit{expert augmentation}. Based on a probabilistic formalization of hybrid modelling, we show why expert augmentation improves generalization. Finally, we validate the practical benefits of augmented hybrid models on a set of controlled experiments, modelling dynamical systems described by ordinary and partial differential equations.
Learning Invariant Representations with Missing Data
Dec 01, 2021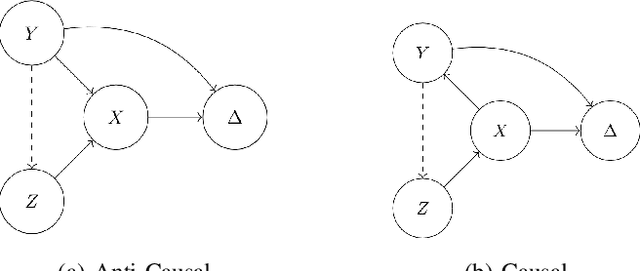
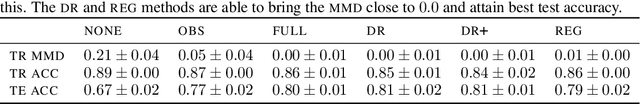


Abstract:Spurious correlations allow flexible models to predict well during training but poorly on related test populations. Recent work has shown that models that satisfy particular independencies involving correlation-inducing \textit{nuisance} variables have guarantees on their test performance. Enforcing such independencies requires nuisances to be observed during training. However, nuisances, such as demographics or image background labels, are often missing. Enforcing independence on just the observed data does not imply independence on the entire population. Here we derive \acrshort{mmd} estimators used for invariance objectives under missing nuisances. On simulations and clinical data, optimizing through these estimates achieves test performance similar to using estimators that make use of the full data.
Exchanging Lessons Between Algorithmic Fairness and Domain Generalization
Oct 14, 2020



Abstract:Standard learning approaches are designed to perform well on average for the data distribution available at training time. Developing learning approaches that are not overly sensitive to the training distribution is central to research on domain- or out-of-distribution generalization, robust optimization and fairness. In this work we focus on links between research on domain generalization and algorithmic fairness -- where performance under a distinct but related test distributions is studied -- and show how the two fields can be mutually beneficial. While domain generalization methods typically rely on knowledge of disjoint "domains" or "environments", "sensitive" label information indicating which demographic groups are at risk of discrimination is often used in the fairness literature. Drawing inspiration from recent fairness approaches that improve worst-case performance without knowledge of sensitive groups, we propose a novel domain generalization method that handles the more realistic scenario where environment partitions are not provided. We then show theoretically and empirically how different partitioning schemes can lead to increased or decreased generalization performance, enabling us to outperform Invariant Risk Minimization with handcrafted environments in multiple cases. We also show how a re-interpretation of IRMv1 allows us for the first time to directly optimize a common fairness criterion, group-sufficiency, and thereby improve performance on a fair prediction task.
Understanding and mitigating exploding inverses in invertible neural networks
Jun 16, 2020
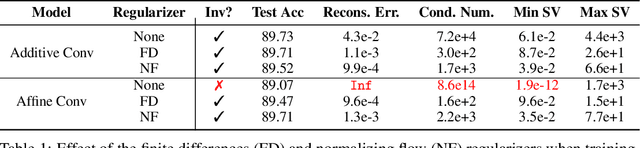
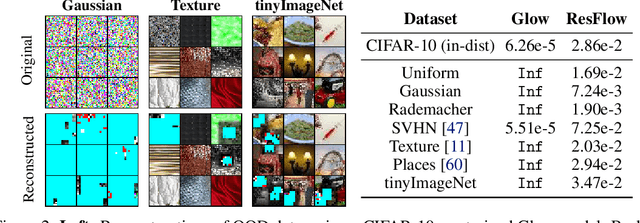
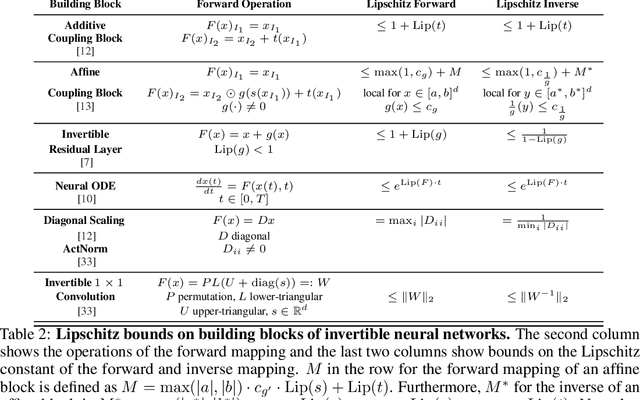
Abstract:Invertible neural networks (INNs) have been used to design generative models, implement memory-saving gradient computation, and solve inverse problems. In this work, we show that commonly-used INN architectures suffer from exploding inverses and are thus prone to becoming numerically non-invertible. Across a wide range of INN use-cases, we reveal failures including the non-applicability of the change-of-variables formula on in- and out-of-distribution (OOD) data, incorrect gradients for memory-saving backprop, and the inability to sample from normalizing flow models. We further derive bi-Lipschitz properties of atomic building blocks of common architectures. These insights into the stability of INNs then provide ways forward to remedy these failures. For tasks where local invertibility is sufficient, like memory-saving backprop, we propose a flexible and efficient regularizer. For problems where global invertibility is necessary, such as applying normalizing flows on OOD data, we show the importance of designing stable INN building blocks.
Shortcut Learning in Deep Neural Networks
May 20, 2020


Abstract:Deep learning has triggered the current rise of artificial intelligence and is the workhorse of today's machine intelligence. Numerous success stories have rapidly spread all over science, industry and society, but its limitations have only recently come into focus. In this perspective we seek to distil how many of deep learning's problem can be seen as different symptoms of the same underlying problem: shortcut learning. Shortcuts are decision rules that perform well on standard benchmarks but fail to transfer to more challenging testing conditions, such as real-world scenarios. Related issues are known in Comparative Psychology, Education and Linguistics, suggesting that shortcut learning may be a common characteristic of learning systems, biological and artificial alike. Based on these observations, we develop a set of recommendations for model interpretation and benchmarking, highlighting recent advances in machine learning to improve robustness and transferability from the lab to real-world applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge