Considerations for Distribution Shift Robustness of Diagnostic Models in Healthcare
Paper and Code
Oct 25, 2024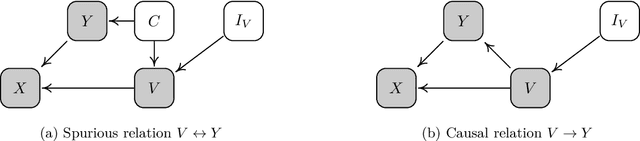
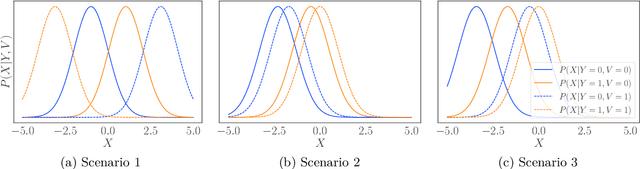
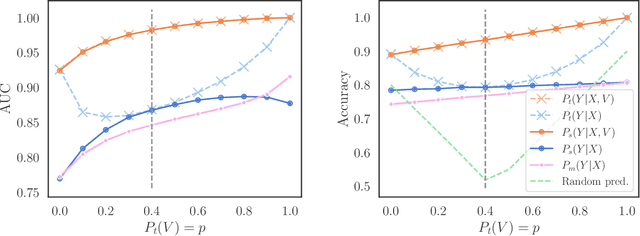
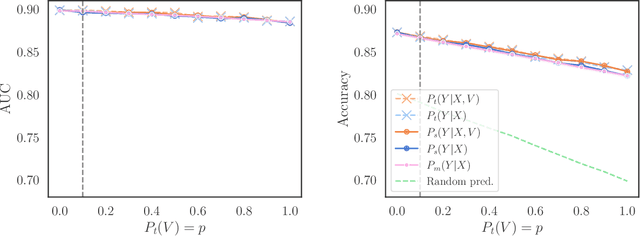
We consider robustness to distribution shifts in the context of diagnostic models in healthcare, where the prediction target $Y$, e.g., the presence of a disease, is causally upstream of the observations $X$, e.g., a biomarker. Distribution shifts may occur, for instance, when the training data is collected in a domain with patients having particular demographic characteristics while the model is deployed on patients from a different demographic group. In the domain of applied ML for health, it is common to predict $Y$ from $X$ without considering further information about the patient. However, beyond the direct influence of the disease $Y$ on biomarker $X$, a predictive model may learn to exploit confounding dependencies (or shortcuts) between $X$ and $Y$ that are unstable under certain distribution shifts. In this work, we highlight a data generating mechanism common to healthcare settings and discuss how recent theoretical results from the causality literature can be applied to build robust predictive models. We theoretically show why ignoring covariates as well as common invariant learning approaches will in general not yield robust predictors in the studied setting, while including certain covariates into the prediction model will. In an extensive simulation study, we showcase the robustness (or lack thereof) of different predictors under various data generating processes. Lastly, we analyze the performance of the different approaches using the PTB-XL dataset, a public dataset of annotated ECG recordings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge