Adam Goliński
Trained on Tokens, Calibrated on Concepts: The Emergence of Semantic Calibration in LLMs
Nov 06, 2025Abstract:Large Language Models (LLMs) often lack meaningful confidence estimates for their outputs. While base LLMs are known to exhibit next-token calibration, it remains unclear whether they can assess confidence in the actual meaning of their responses beyond the token level. We find that, when using a certain sampling-based notion of semantic calibration, base LLMs are remarkably well-calibrated: they can meaningfully assess confidence in open-domain question-answering tasks, despite not being explicitly trained to do so. Our main theoretical contribution establishes a mechanism for why semantic calibration emerges as a byproduct of next-token prediction, leveraging a recent connection between calibration and local loss optimality. The theory relies on a general definition of "B-calibration," which is a notion of calibration parameterized by a choice of equivalence classes (semantic or otherwise). This theoretical mechanism leads to a testable prediction: base LLMs will be semantically calibrated when they can easily predict their own distribution over semantic answer classes before generating a response. We state three implications of this prediction, which we validate through experiments: (1) Base LLMs are semantically calibrated across question-answering tasks, (2) RL instruction-tuning systematically breaks this calibration, and (3) chain-of-thought reasoning breaks calibration. To our knowledge, our work provides the first principled explanation of when and why semantic calibration emerges in LLMs.
BED-LLM: Intelligent Information Gathering with LLMs and Bayesian Experimental Design
Aug 28, 2025Abstract:We propose a general-purpose approach for improving the ability of Large Language Models (LLMs) to intelligently and adaptively gather information from a user or other external source using the framework of sequential Bayesian experimental design (BED). This enables LLMs to act as effective multi-turn conversational agents and interactively interface with external environments. Our approach, which we call BED-LLM (Bayesian Experimental Design with Large Language Models), is based on iteratively choosing questions or queries that maximize the expected information gain (EIG) about the task of interest given the responses gathered previously. We show how this EIG can be formulated in a principled way using a probabilistic model derived from the LLM's belief distribution and provide detailed insights into key decisions in its construction. Further key to the success of BED-LLM are a number of specific innovations, such as a carefully designed estimator for the EIG, not solely relying on in-context updates for conditioning on previous responses, and a targeted strategy for proposing candidate queries. We find that BED-LLM achieves substantial gains in performance across a wide range of tests based on the 20-questions game and using the LLM to actively infer user preferences, compared to direct prompting of the LLM and other adaptive design strategies.
Self-reflective Uncertainties: Do LLMs Know Their Internal Answer Distribution?
May 26, 2025Abstract:To reveal when a large language model (LLM) is uncertain about a response, uncertainty quantification commonly produces percentage numbers along with the output. But is this all we can do? We argue that in the output space of LLMs, the space of strings, exist strings expressive enough to summarize the distribution over output strings the LLM deems possible. We lay a foundation for this new avenue of uncertainty explication and present SelfReflect, a theoretically-motivated metric to assess how faithfully a string summarizes an LLM's internal answer distribution. We show that SelfReflect is able to discriminate even subtle differences of candidate summary strings and that it aligns with human judgement, outperforming alternative metrics such as LLM judges and embedding comparisons. With SelfReflect, we investigate a number of self-summarization methods and find that even state-of-the-art reasoning models struggle to explicate their internal uncertainty. But we find that faithful summarizations can be generated by sampling and summarizing. Our metric enables future works towards this universal form of LLM uncertainties.
Considerations for Distribution Shift Robustness of Diagnostic Models in Healthcare
Oct 25, 2024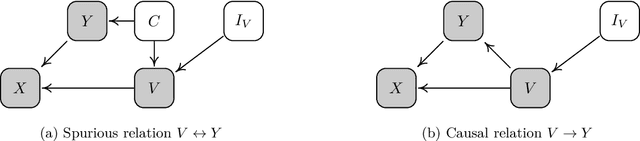
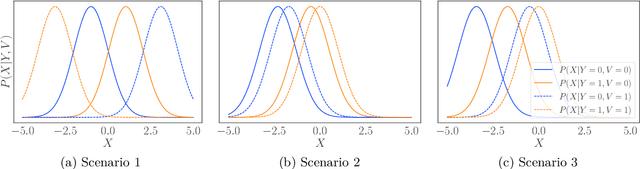
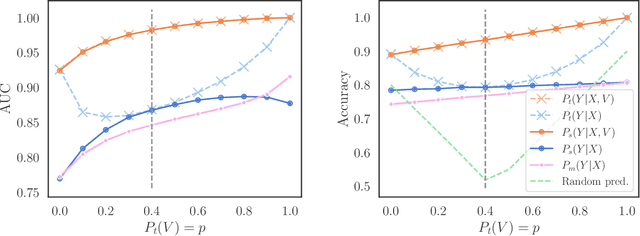
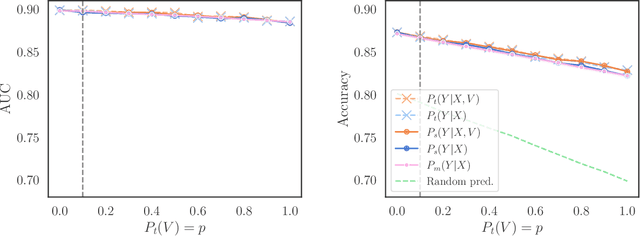
Abstract:We consider robustness to distribution shifts in the context of diagnostic models in healthcare, where the prediction target $Y$, e.g., the presence of a disease, is causally upstream of the observations $X$, e.g., a biomarker. Distribution shifts may occur, for instance, when the training data is collected in a domain with patients having particular demographic characteristics while the model is deployed on patients from a different demographic group. In the domain of applied ML for health, it is common to predict $Y$ from $X$ without considering further information about the patient. However, beyond the direct influence of the disease $Y$ on biomarker $X$, a predictive model may learn to exploit confounding dependencies (or shortcuts) between $X$ and $Y$ that are unstable under certain distribution shifts. In this work, we highlight a data generating mechanism common to healthcare settings and discuss how recent theoretical results from the causality literature can be applied to build robust predictive models. We theoretically show why ignoring covariates as well as common invariant learning approaches will in general not yield robust predictors in the studied setting, while including certain covariates into the prediction model will. In an extensive simulation study, we showcase the robustness (or lack thereof) of different predictors under various data generating processes. Lastly, we analyze the performance of the different approaches using the PTB-XL dataset, a public dataset of annotated ECG recordings.
The Role of Entropy and Reconstruction in Multi-View Self-Supervised Learning
Jul 20, 2023



Abstract:The mechanisms behind the success of multi-view self-supervised learning (MVSSL) are not yet fully understood. Contrastive MVSSL methods have been studied through the lens of InfoNCE, a lower bound of the Mutual Information (MI). However, the relation between other MVSSL methods and MI remains unclear. We consider a different lower bound on the MI consisting of an entropy and a reconstruction term (ER), and analyze the main MVSSL families through its lens. Through this ER bound, we show that clustering-based methods such as DeepCluster and SwAV maximize the MI. We also re-interpret the mechanisms of distillation-based approaches such as BYOL and DINO, showing that they explicitly maximize the reconstruction term and implicitly encourage a stable entropy, and we confirm this empirically. We show that replacing the objectives of common MVSSL methods with this ER bound achieves competitive performance, while making them stable when training with smaller batch sizes or smaller exponential moving average (EMA) coefficients. Github repo: https://github.com/apple/ml-entropy-reconstruction.
LIDL: Local Intrinsic Dimension Estimation Using Approximate Likelihood
Jun 29, 2022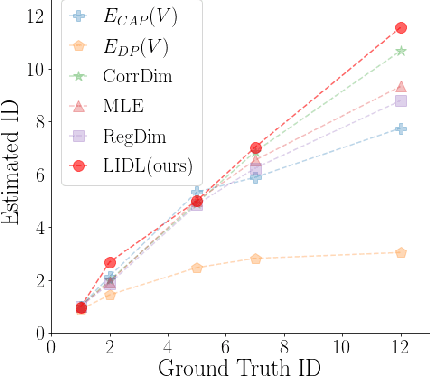
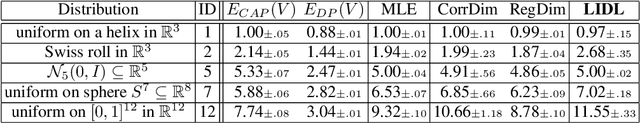
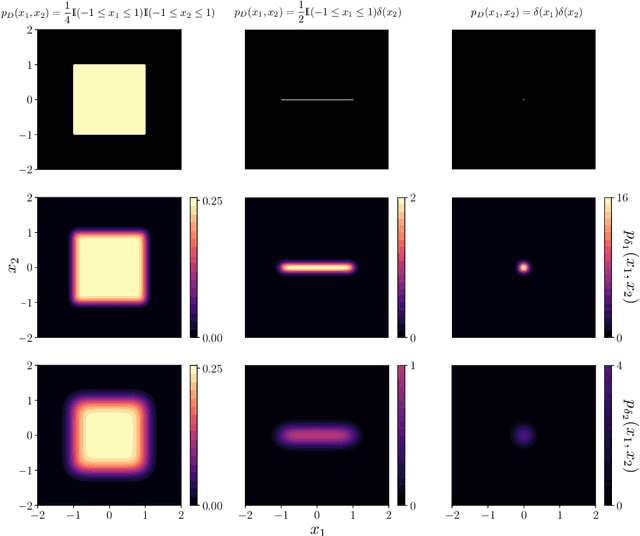
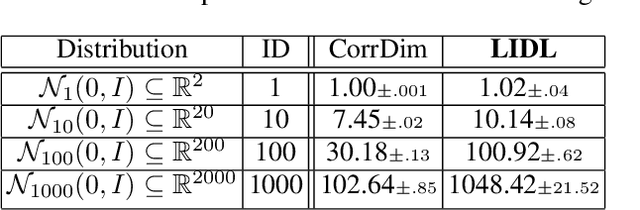
Abstract:Most of the existing methods for estimating the local intrinsic dimension of a data distribution do not scale well to high-dimensional data. Many of them rely on a non-parametric nearest neighbors approach which suffers from the curse of dimensionality. We attempt to address that challenge by proposing a novel approach to the problem: Local Intrinsic Dimension estimation using approximate Likelihood (LIDL). Our method relies on an arbitrary density estimation method as its subroutine and hence tries to sidestep the dimensionality challenge by making use of the recent progress in parametric neural methods for likelihood estimation. We carefully investigate the empirical properties of the proposed method, compare them with our theoretical predictions, and show that LIDL yields competitive results on the standard benchmarks for this problem and that it scales to thousands of dimensions. What is more, we anticipate this approach to improve further with the continuing advances in the density estimation literature.
COIN++: Data Agnostic Neural Compression
Jan 30, 2022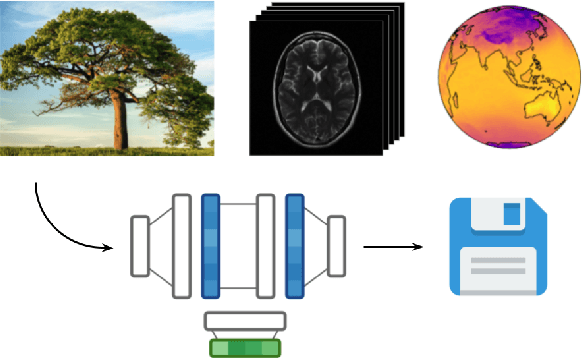
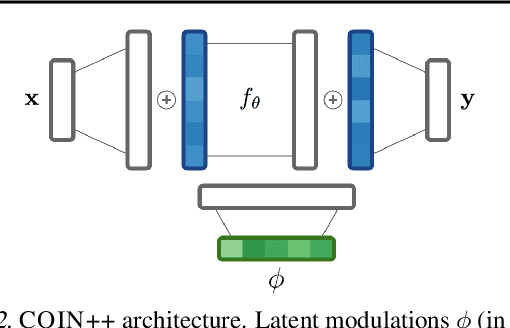
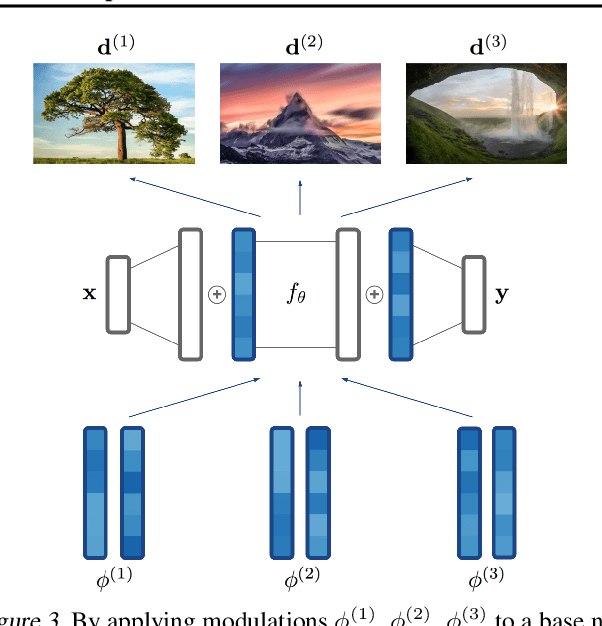
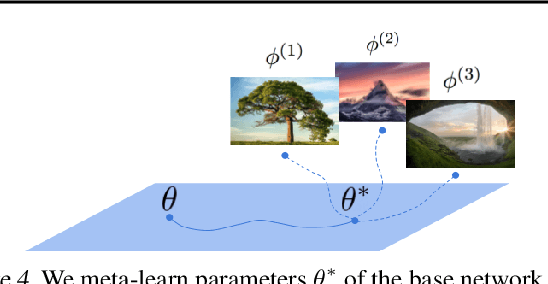
Abstract:Neural compression algorithms are typically based on autoencoders that require specialized encoder and decoder architectures for different data modalities. In this paper, we propose COIN++, a neural compression framework that seamlessly handles a wide range of data modalities. Our approach is based on converting data to implicit neural representations, i.e. neural functions that map coordinates (such as pixel locations) to features (such as RGB values). Then, instead of storing the weights of the implicit neural representation directly, we store modulations applied to a meta-learned base network as a compressed code for the data. We further quantize and entropy code these modulations, leading to large compression gains while reducing encoding time by two orders of magnitude compared to baselines. We empirically demonstrate the effectiveness of our method by compressing various data modalities, from images to medical and climate data.
Expectation Programming
Jun 09, 2021



Abstract:Building on ideas from probabilistic programming, we introduce the concept of an expectation programming framework (EPF) that automates the calculation of expectations. Analogous to a probabilistic program, an expectation program is comprised of a mix of probabilistic constructs and deterministic calculations that define a conditional distribution over its variables. However, the focus of the inference engine in an EPF is to directly estimate the resulting expectation of the program return values, rather than approximate the conditional distribution itself. This distinction allows us to achieve substantial performance improvements over the standard probabilistic programming pipeline by tailoring the inference to the precise expectation we care about. We realize a particular instantiation of our EPF concept by extending the probabilistic programming language Turing to allow so-called target-aware inference to be run automatically, and show that this leads to significant empirical gains compared to conventional posterior-based inference.
COIN: COmpression with Implicit Neural representations
Mar 03, 2021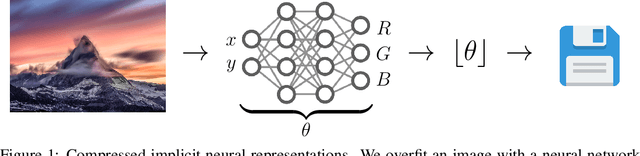
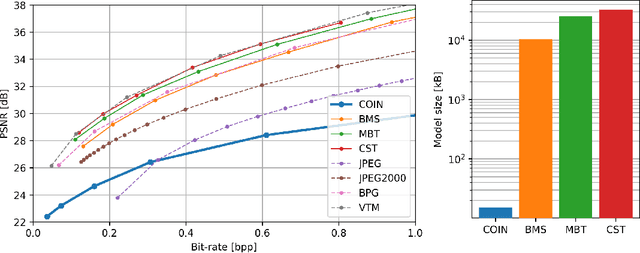
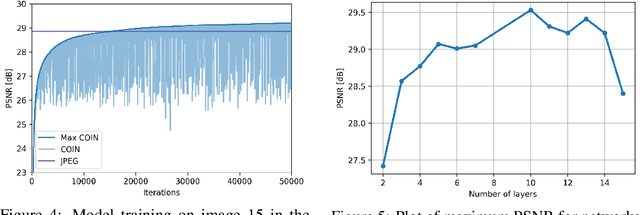
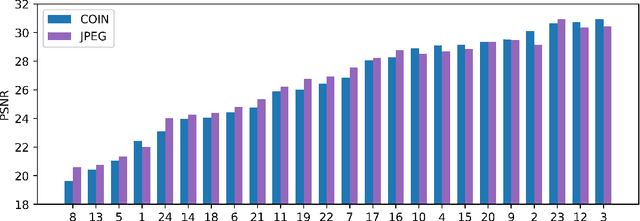
Abstract:We propose a new simple approach for image compression: instead of storing the RGB values for each pixel of an image, we store the weights of a neural network overfitted to the image. Specifically, to encode an image, we fit it with an MLP which maps pixel locations to RGB values. We then quantize and store the weights of this MLP as a code for the image. To decode the image, we simply evaluate the MLP at every pixel location. We found that this simple approach outperforms JPEG at low bit-rates, even without entropy coding or learning a distribution over weights. While our framework is not yet competitive with state of the art compression methods, we show that it has various attractive properties which could make it a viable alternative to other neural data compression approaches.
Amortized Monte Carlo Integration
Jul 18, 2019



Abstract:Current approaches to amortizing Bayesian inference focus solely on approximating the posterior distribution. Typically, this approximation is, in turn, used to calculate expectations for one or more target functions - a computational pipeline which is inefficient when the target function(s) are known upfront. In this paper, we address this inefficiency by introducing AMCI, a method for amortizing Monte Carlo integration directly. AMCI operates similarly to amortized inference but produces three distinct amortized proposals, each tailored to a different component of the overall expectation calculation. At runtime, samples are produced separately from each amortized proposal, before being combined to an overall estimate of the expectation. We show that while existing approaches are fundamentally limited in the level of accuracy they can achieve, AMCI can theoretically produce arbitrarily small errors for any integrable target function using only a single sample from each proposal at runtime. We further show that it is able to empirically outperform the theoretically optimal self-normalized importance sampler on a number of example problems. Furthermore, AMCI allows not only for amortizing over datasets but also amortizing over target functions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge