Hrushikesh Loya
How Much Is Hidden in the NAS Benchmarks? Few-Shot Adaptation of a NAS Predictor
Nov 30, 2023
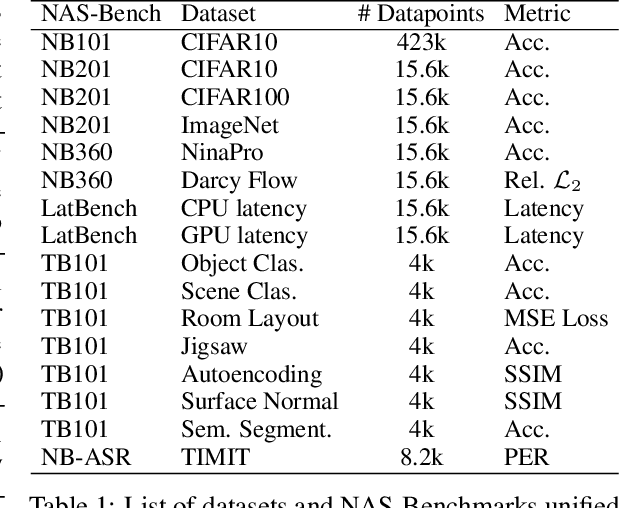
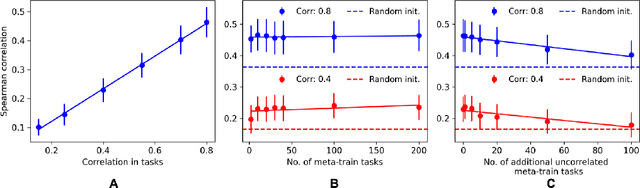
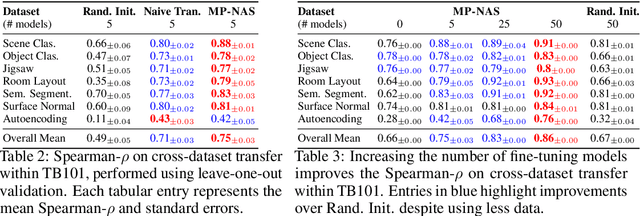
Abstract:Neural architecture search has proven to be a powerful approach to designing and refining neural networks, often boosting their performance and efficiency over manually-designed variations, but comes with computational overhead. While there has been a considerable amount of research focused on lowering the cost of NAS for mainstream tasks, such as image classification, a lot of those improvements stem from the fact that those tasks are well-studied in the broader context. Consequently, applicability of NAS to emerging and under-represented domains is still associated with a relatively high cost and/or uncertainty about the achievable gains. To address this issue, we turn our focus towards the recent growth of publicly available NAS benchmarks in an attempt to extract general NAS knowledge, transferable across different tasks and search spaces. We borrow from the rich field of meta-learning for few-shot adaptation and carefully study applicability of those methods to NAS, with a special focus on the relationship between task-level correlation (domain shift) and predictor transferability; which we deem critical for improving NAS on diverse tasks. In our experiments, we use 6 NAS benchmarks in conjunction, spanning in total 16 NAS settings -- our meta-learning approach not only shows superior (or matching) performance in the cross-validation experiments but also successful extrapolation to a new search space and tasks.
COIN++: Data Agnostic Neural Compression
Jan 30, 2022
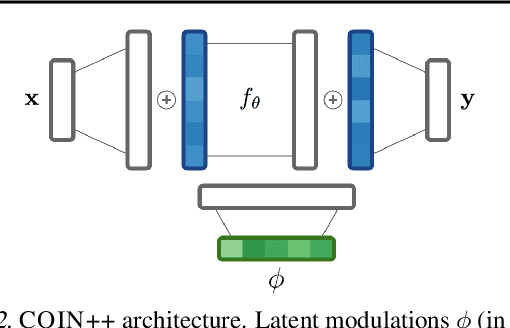
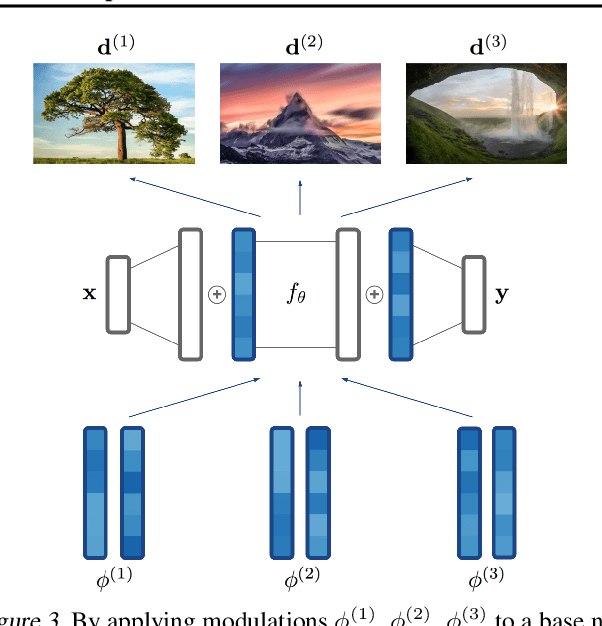
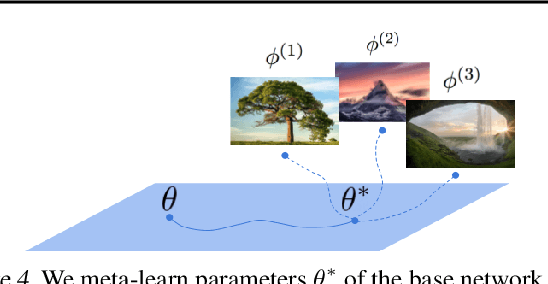
Abstract:Neural compression algorithms are typically based on autoencoders that require specialized encoder and decoder architectures for different data modalities. In this paper, we propose COIN++, a neural compression framework that seamlessly handles a wide range of data modalities. Our approach is based on converting data to implicit neural representations, i.e. neural functions that map coordinates (such as pixel locations) to features (such as RGB values). Then, instead of storing the weights of the implicit neural representation directly, we store modulations applied to a meta-learned base network as a compressed code for the data. We further quantize and entropy code these modulations, leading to large compression gains while reducing encoding time by two orders of magnitude compared to baselines. We empirically demonstrate the effectiveness of our method by compressing various data modalities, from images to medical and climate data.
Functional Space Variational Inference for Uncertainty Estimation in Computer Aided Diagnosis
May 28, 2020
Abstract:Deep neural networks have revolutionized medical image analysis and disease diagnosis. Despite their impressive performance, it is difficult to generate well-calibrated probabilistic outputs for such networks, which makes them uninterpretable black boxes. Bayesian neural networks provide a principled approach for modelling uncertainty and increasing patient safety, but they have a large computational overhead and provide limited improvement in calibration. In this work, by taking skin lesion classification as an example task, we show that by shifting Bayesian inference to the functional space we can craft meaningful priors that give better calibrated uncertainty estimates at a much lower computational cost.
* Meaningful priors on the functional space rather than the weight space, result in well calibrated uncertainty estimates
Uncertainty Estimation in Cancer Survival Prediction
Mar 25, 2020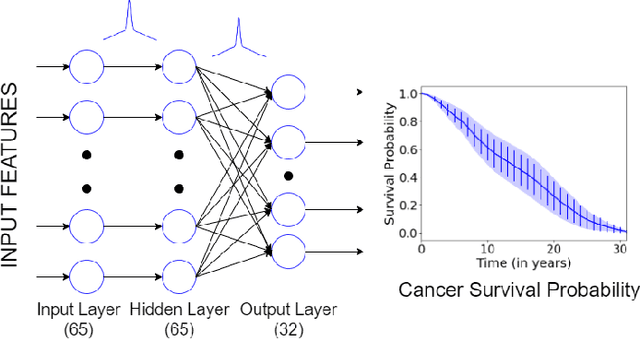
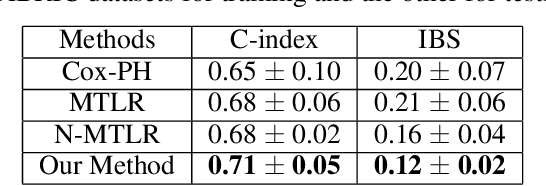
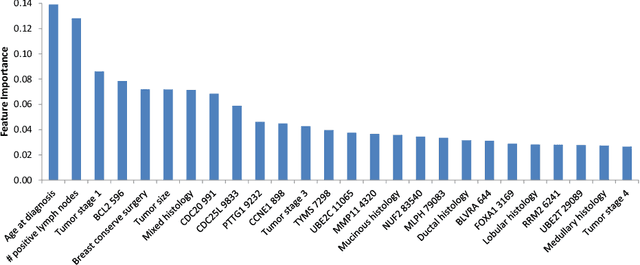

Abstract:Survival models are used in various fields, such as the development of cancer treatment protocols. Although many statistical and machine learning models have been proposed to achieve accurate survival predictions, little attention has been paid to obtain well-calibrated uncertainty estimates associated with each prediction. The currently popular models are opaque and untrustworthy in that they often express high confidence even on those test cases that are not similar to the training samples, and even when their predictions are wrong. We propose a Bayesian framework for survival models that not only gives more accurate survival predictions but also quantifies the survival uncertainty better. Our approach is a novel combination of variational inference for uncertainty estimation, neural multi-task logistic regression for estimating nonlinear and time-varying risk models, and an additional sparsity-inducing prior to work with high dimensional data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge