Deepak Anand
Switching Loss for Generalized Nucleus Detection in Histopathology
Aug 09, 2020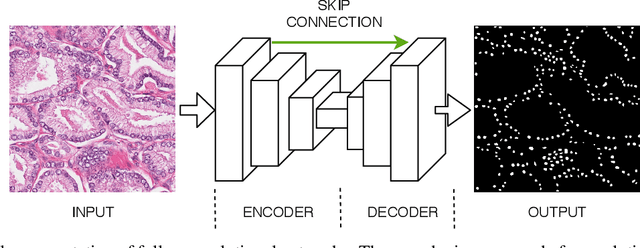
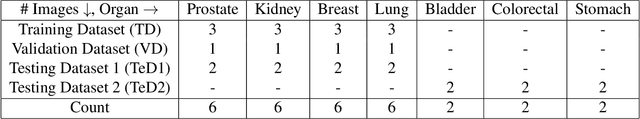
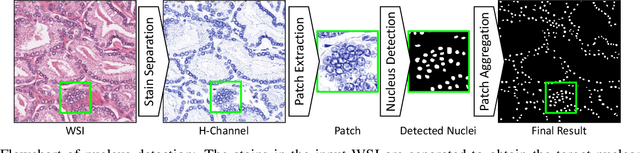
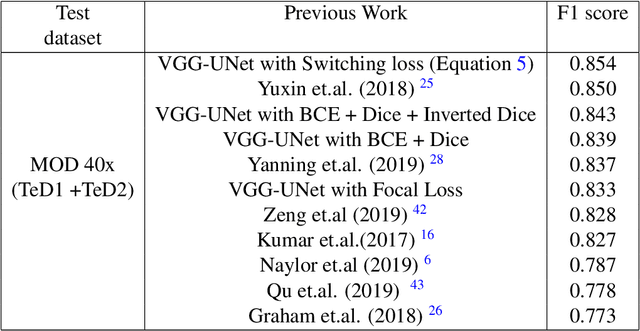
Abstract:The accuracy of deep learning methods for two foundational tasks in medical image analysis -- detection and segmentation -- can suffer from class imbalance. We propose a `switching loss' function that adaptively shifts the emphasis between foreground and background classes. While the existing loss functions to address this problem were motivated by the classification task, the switching loss is based on Dice loss, which is better suited for segmentation and detection. Furthermore, to get the most out the training samples, we adapt the loss with each mini-batch, unlike previous proposals that adapt once for the entire training set. A nucleus detector trained using the proposed loss function on a source dataset outperformed those trained using cross-entropy, Dice, or focal losses. Remarkably, without retraining on target datasets, our pre-trained nucleus detector also outperformed existing nucleus detectors that were trained on at least some of the images from the target datasets. To establish a broad utility of the proposed loss, we also confirmed that it led to more accurate ventricle segmentation in MRI as compared to the other loss functions. Our GPU-enabled pre-trained nucleus detection software is also ready to process whole slide images right out-of-the-box and is usably fast.
Visualization for Histopathology Images using Graph Convolutional Neural Networks
Jun 16, 2020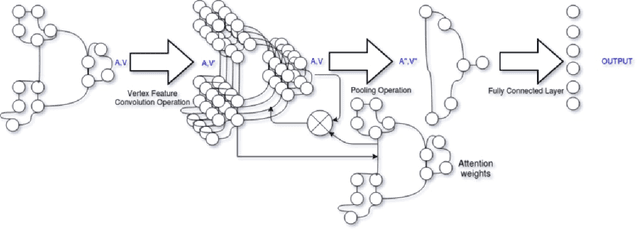
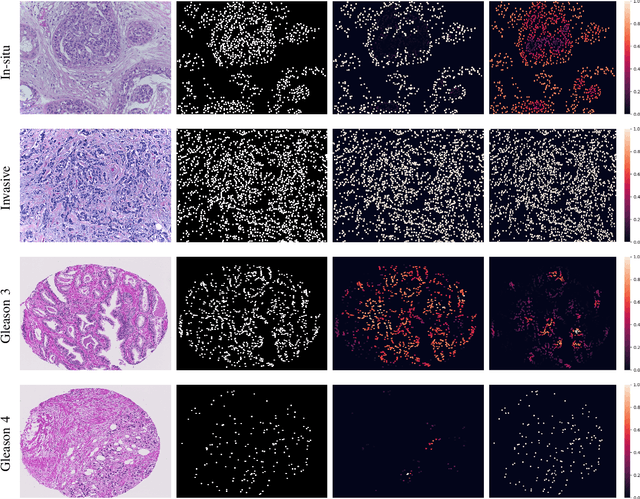
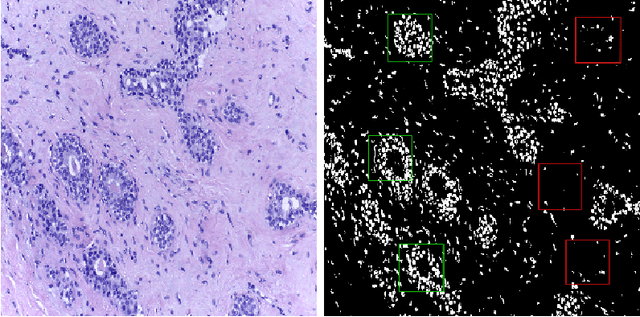
Abstract:With the increase in the use of deep learning for computer-aided diagnosis in medical images, the criticism of the black-box nature of the deep learning models is also on the rise. The medical community needs interpretable models for both due diligence and advancing the understanding of disease and treatment mechanisms. In histology, in particular, while there is rich detail available at the cellular level and that of spatial relationships between cells, it is difficult to modify convolutional neural networks to point out the relevant visual features. We adopt an approach to model histology tissue as a graph of nuclei and develop a graph convolutional network framework based on attention mechanism and node occlusion for disease diagnosis. The proposed method highlights the relative contribution of each cell nucleus in the whole-slide image. Our visualization of such networks trained to distinguish between invasive and in-situ breast cancers, and Gleason 3 and 4 prostate cancers generate interpretable visual maps that correspond well with our understanding of the structures that are important to experts for their diagnosis.
Uncertainty Estimation in Cancer Survival Prediction
Mar 25, 2020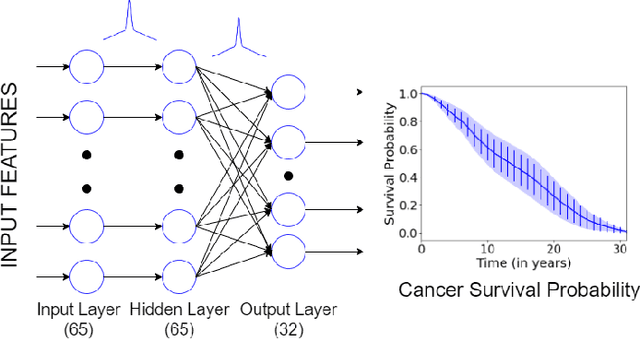
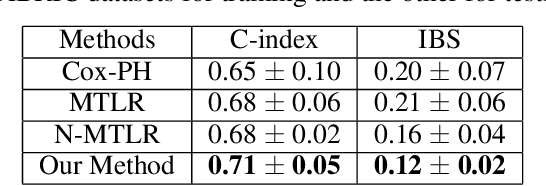
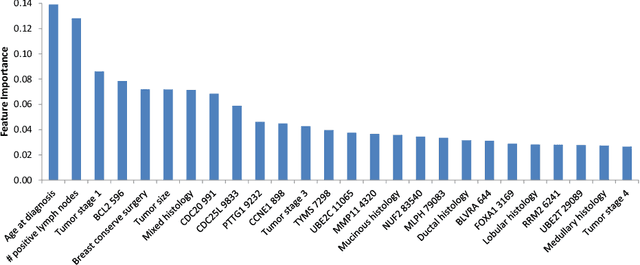

Abstract:Survival models are used in various fields, such as the development of cancer treatment protocols. Although many statistical and machine learning models have been proposed to achieve accurate survival predictions, little attention has been paid to obtain well-calibrated uncertainty estimates associated with each prediction. The currently popular models are opaque and untrustworthy in that they often express high confidence even on those test cases that are not similar to the training samples, and even when their predictions are wrong. We propose a Bayesian framework for survival models that not only gives more accurate survival predictions but also quantifies the survival uncertainty better. Our approach is a novel combination of variational inference for uncertainty estimation, neural multi-task logistic regression for estimating nonlinear and time-varying risk models, and an additional sparsity-inducing prior to work with high dimensional data.
Breast Cancer Histopathology Image Classification and Localization using Multiple Instance Learning
Feb 16, 2020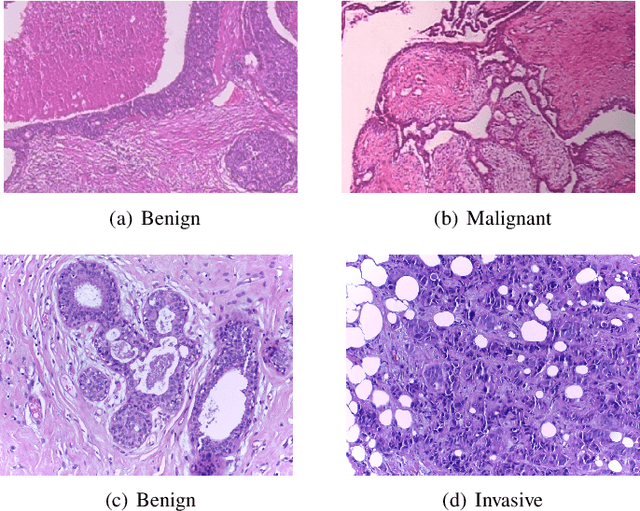
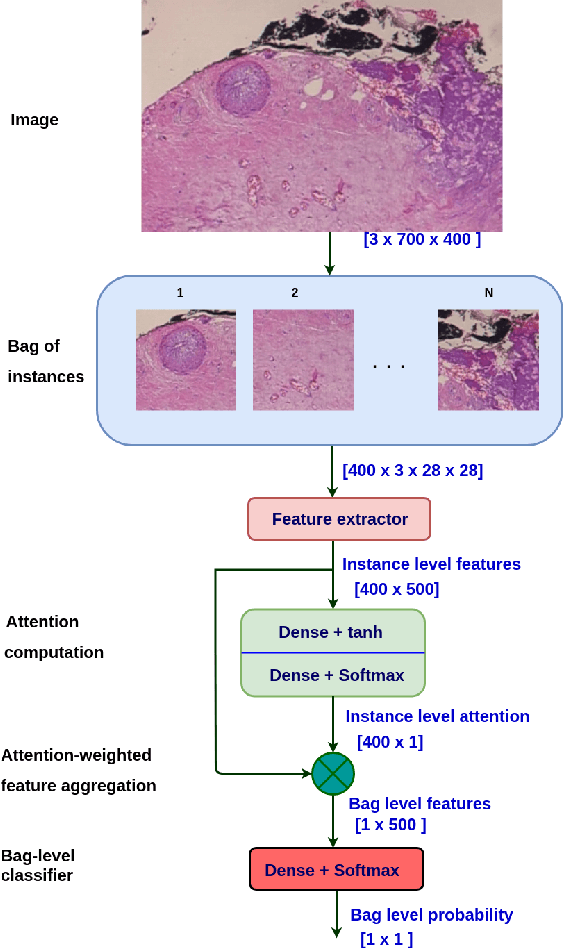
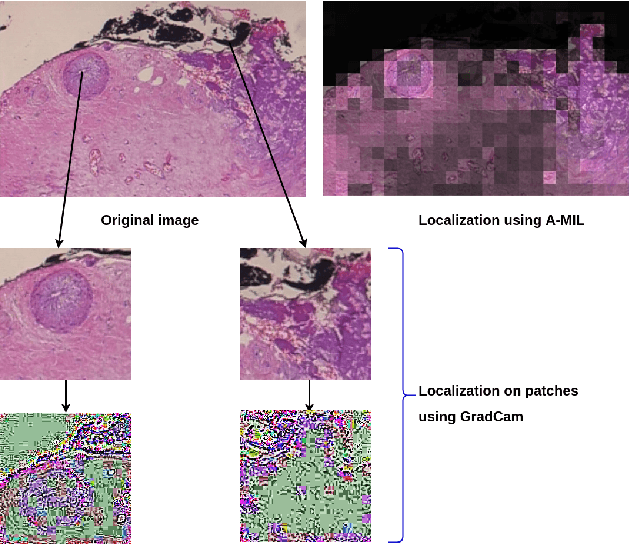
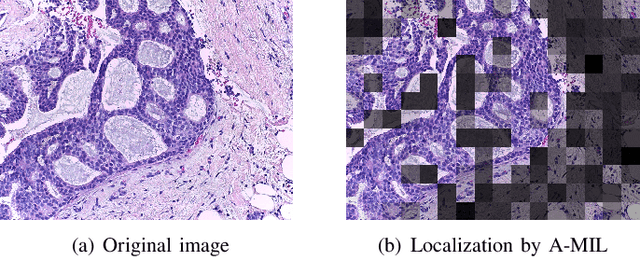
Abstract:Breast cancer has the highest mortality among cancers in women. Computer-aided pathology to analyze microscopic histopathology images for diagnosis with an increasing number of breast cancer patients can bring the cost and delays of diagnosis down. Deep learning in histopathology has attracted attention over the last decade of achieving state-of-the-art performance in classification and localization tasks. The convolutional neural network, a deep learning framework, provides remarkable results in tissue images analysis, but lacks in providing interpretation and reasoning behind the decisions. We aim to provide a better interpretation of classification results by providing localization on microscopic histopathology images. We frame the image classification problem as weakly supervised multiple instance learning problem where an image is collection of patches i.e. instances. Attention-based multiple instance learning (A-MIL) learns attention on the patches from the image to localize the malignant and normal regions in an image and use them to classify the image. We present classification and localization results on two publicly available BreakHIS and BACH dataset. The classification and visualization results are compared with other recent techniques. The proposed method achieves better localization results without compromising classification accuracy.
Pixel-wise Segmentation of Right Ventricle of Heart
Aug 21, 2019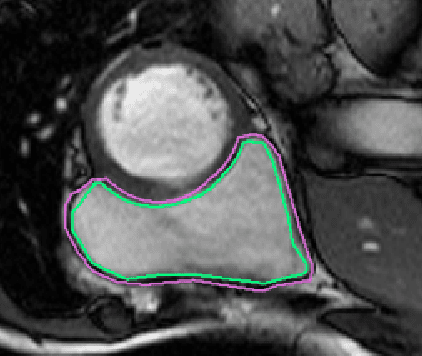
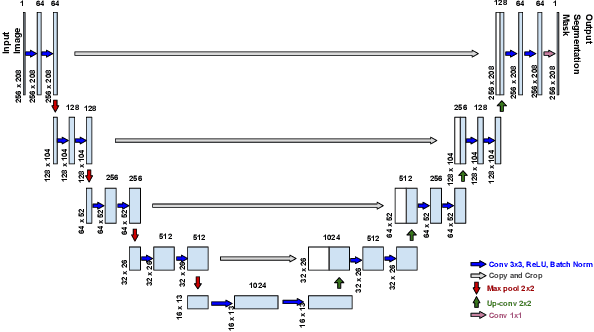
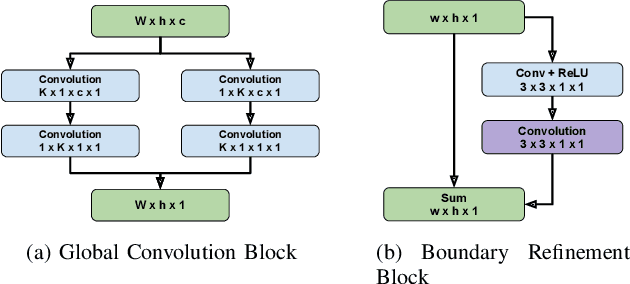
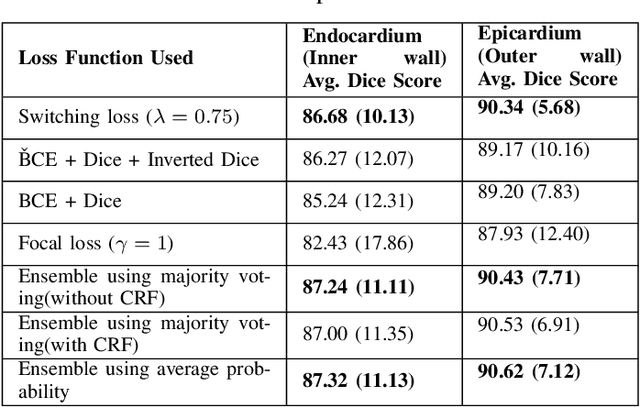
Abstract:One of the first steps in the diagnosis of most cardiac diseases, such as pulmonary hypertension, coronary heart disease is the segmentation of ventricles from cardiac magnetic resonance (MRI) images. Manual segmentation of the right ventricle requires diligence and time, while its automated segmentation is challenging due to shape variations and illdefined borders. We propose a deep learning based method for the accurate segmentation of right ventricle, which does not require post-processing and yet it achieves the state-of-the-art performance of 0.86 Dice coefficient and 6.73 mm Hausdorff distance on RVSC-MICCAI 2012 dataset. We use a novel adaptive cost function to counter extreme class-imbalance in the dataset. We present a comprehensive comparative study of loss functions, architectures, and ensembling techniques to build a principled approach for biomedical segmentation tasks.
Histographs: Graphs in Histopathology
Aug 14, 2019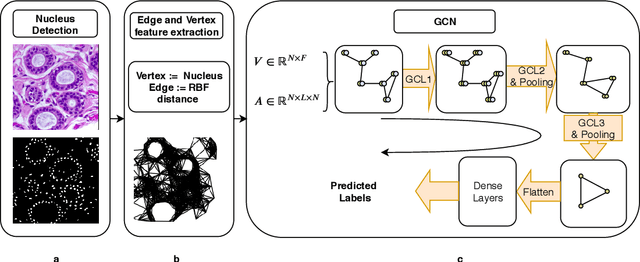
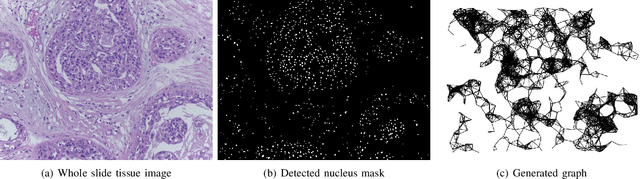
Abstract:Spatial arrangement of cells of various types, such as tumor infiltrating lymphocytes and the advancing edge of a tumor, are important features for detecting and characterizing cancers. However, convolutional neural networks (CNNs) do not explicitly extract intricate features of the spatial arrangements of the cells from histopathology images. In this work, we propose to classify cancers using graph convolutional networks (GCNs) by modeling a tissue section as a multi-attributed spatial graph of its constituent cells. Cells are detected using their nuclei in H&E stained tissue image, and each cell's appearance is captured as a multi-attributed high-dimensional vertex feature. The spatial relations between neighboring cells are captured as edge features based on their distances in a graph. We demonstrate the utility of this approach by obtaining classification accuracy that is competitive with CNNs, specifically, Inception-v3, on two tasks-cancerous versus non-cancerous and in situ versus invasive-on the BACH breast cancer dataset.
MIST: A Novel Training Strategy for Low-latencyScalable Neural Net Decoders
May 22, 2019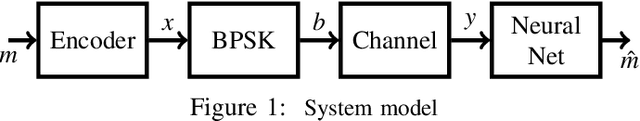
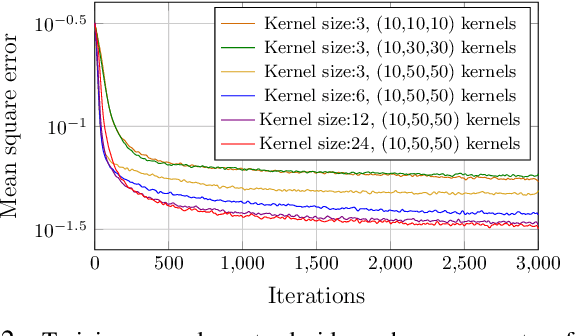


Abstract:In this paper, we propose a low latency, robust and scalable neural net based decoder for convolutional and low-density parity-check (LPDC) coding schemes. The proposed decoders are demonstrated to have bit error rate (BER) and block error rate (BLER) performances at par with the state-of-the-art neural net based decoders while achieving more than 8 times higher decoding speed. The enhanced decoding speed is due to the use of convolutional neural network (CNN) as opposed to recurrent neural network (RNN) used in the best known neural net based decoders. This contradicts existing doctrine that only RNN based decoders can provide a performance close to the optimal ones. The key ingredient to our approach is a novel Mixed-SNR Independent Samples based Training (MIST), which allows for training of CNN with only 1\% of possible datawords, even for block length as high as 1000. The proposed decoder is robust as, once trained, the same decoder can be used for a wide range of SNR values. Finally, in the presence of channel outages, the proposed decoders outperform the best known decoders, {\it viz.} unquantized Viterbi decoder for convolutional code, and belief propagation for LDPC. This gives the CNN decoder a significant advantage in 5G millimeter wave systems, where channel outages are prevalent.
Fast GPU-Enabled Color Normalization for Digital Pathology
Jan 10, 2019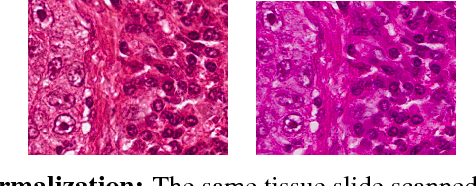
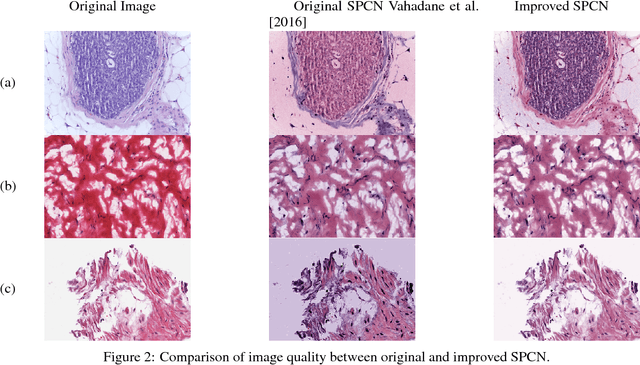

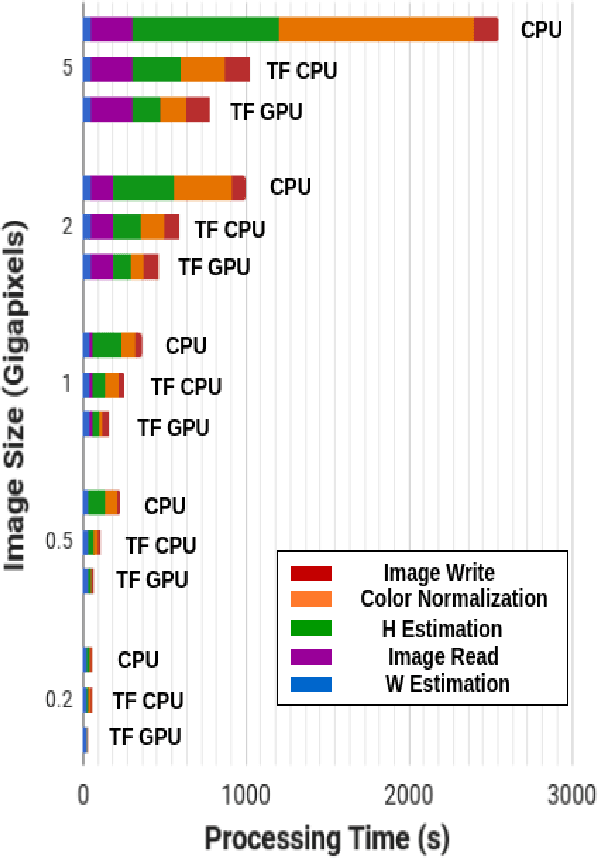
Abstract:Normalizing unwanted color variations due to differences in staining processes and scanner responses has been shown to aid machine learning in computational pathology. Of the several popular techniques for color normalization, structure preserving color normalization (SPCN) is well-motivated, convincingly tested, and published with its code base. However, SPCN makes occasional errors in color basis estimation leading to artifacts such as swapping the color basis vectors between stains or giving a colored tinge to the background with no tissue. We made several algorithmic improvements to remove these artifacts. Additionally, the original SPCN code is not readily usable on gigapixel whole slide images (WSIs) due to long run times, use of proprietary software platform and libraries, and its inability to automatically handle WSIs. We completely rewrote the software such that it can automatically handle images of any size in popular WSI formats. Our software utilizes GPU-acceleration and open-source libraries that are becoming ubiquitous with the advent of deep learning. We also made several other small improvements and achieved a multifold overall speedup on gigapixel images. Our algorithm and software is usable right out-of-the-box by the computational pathology community.
Some New Layer Architectures for Graph CNN
Oct 31, 2018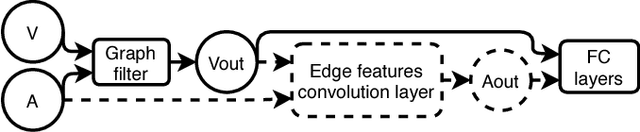



Abstract:While convolutional neural networks (CNNs) have recently made great strides in supervised classification of data structured on a grid (e.g. images composed of pixel grids), in several interesting datasets, the relations between features can be better represented as a general graph instead of a regular grid. Although recent algorithms that adapt CNNs to graphs have shown promising results, they mostly neglect learning explicit operations for edge features while focusing on vertex features alone. We propose new formulations for convolutional, pooling, and fully connected layers for neural networks that make more comprehensive use of the information available in multi-dimensional graphs. Using these layers led to an improvement in classification accuracy over the state-of-the-art methods on benchmark graph datasets.
Classification of Breast Cancer Histology using Deep Learning
Jul 25, 2018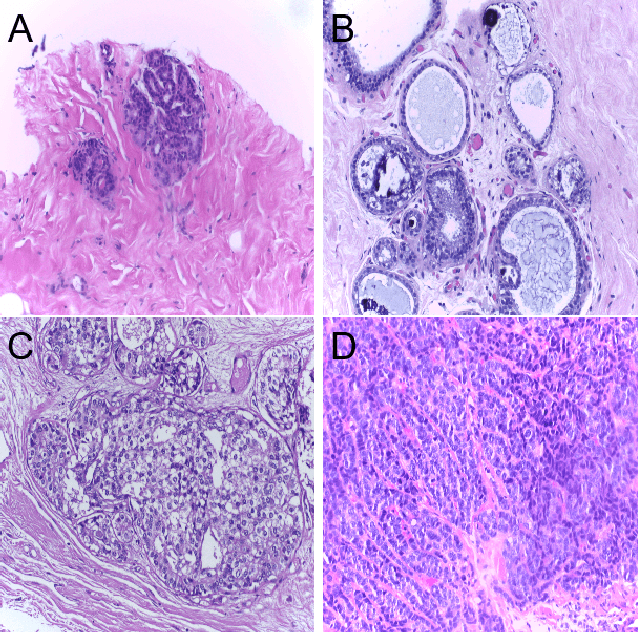

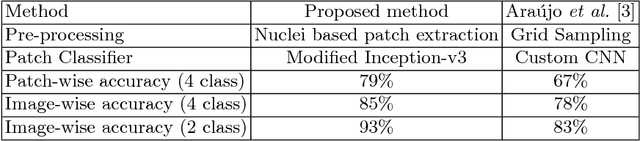
Abstract:Breast Cancer is a major cause of death worldwide among women. Hematoxylin and Eosin (H&E) stained breast tissue samples from biopsies are observed under microscopes for the primary diagnosis of breast cancer. In this paper, we propose a deep learning-based method for classification of H&E stained breast tissue images released for BACH challenge 2018 by fine-tuning Inception-v3 convolutional neural network (CNN) proposed by Szegedy et al. These images are to be classified into four classes namely, i) normal tissue, ii) benign tumor, iii) in-situ carcinoma and iv) invasive carcinoma. Our strategy is to extract patches based on nuclei density instead of random or grid sampling, along with rejection of patches that are not rich in nuclei (non-epithelial) regions for training and testing. Every patch (nuclei-dense region) in an image is classified in one of the four above mentioned categories. The class of the entire image is determined using majority voting over the nuclear classes. We obtained an average four class accuracy of 85% and an average two class (non-cancer vs. carcinoma) accuracy of 93%, which improves upon a previous benchmark by Araujo et al.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge