Amit Sethi
Indian Institute of Technology Bombay
BiPrompt: Bilateral Prompt Optimization for Visual and Textual Debiasing in Vision-Language Models
Jan 05, 2026Abstract:Vision language foundation models such as CLIP exhibit impressive zero-shot generalization yet remain vulnerable to spurious correlations across visual and textual modalities. Existing debiasing approaches often address a single modality either visual or textual leading to partial robustness and unstable adaptation under distribution shifts. We propose a bilateral prompt optimization framework (BiPrompt) that simultaneously mitigates non-causal feature reliance in both modalities during test-time adaptation. On the visual side, it employs structured attention-guided erasure to suppress background activations and enforce orthogonal prediction consistency between causal and spurious regions. On the textual side, it introduces balanced prompt normalization, a learnable re-centering mechanism that aligns class embeddings toward an isotropic semantic space. Together, these modules jointly minimize conditional mutual information between spurious cues and predictions, steering the model toward causal, domain invariant reasoning without retraining or domain supervision. Extensive evaluations on real-world and synthetic bias benchmarks demonstrate consistent improvements in both average and worst-group accuracies over prior test-time debiasing methods, establishing a lightweight yet effective path toward trustworthy and causally grounded vision-language adaptation.
FedHypeVAE: Federated Learning with Hypernetwork Generated Conditional VAEs for Differentially Private Embedding Sharing
Jan 02, 2026Abstract:Federated data sharing promises utility without centralizing raw data, yet existing embedding-level generators struggle under non-IID client heterogeneity and provide limited formal protection against gradient leakage. We propose FedHypeVAE, a differentially private, hypernetwork-driven framework for synthesizing embedding-level data across decentralized clients. Building on a conditional VAE backbone, we replace the single global decoder and fixed latent prior with client-aware decoders and class-conditional priors generated by a shared hypernetwork from private, trainable client codes. This bi-level design personalizes the generative layerrather than the downstream modelwhile decoupling local data from communicated parameters. The shared hypernetwork is optimized under differential privacy, ensuring that only noise-perturbed, clipped gradients are aggregated across clients. A local MMD alignment between real and synthetic embeddings and a Lipschitz regularizer on hypernetwork outputs further enhance stability and distributional coherence under non-IID conditions. After training, a neutral meta-code enables domain agnostic synthesis, while mixtures of meta-codes provide controllable multi-domain coverage. FedHypeVAE unifies personalization, privacy, and distribution alignment at the generator level, establishing a principled foundation for privacy-preserving data synthesis in federated settings. Code: github.com/sunnyinAI/FedHypeVAE
Spatially-Aware Mixture of Experts with Log-Logistic Survival Modeling for Whole-Slide Images
Nov 17, 2025Abstract:Accurate survival prediction from histopathology whole-slide images (WSIs) remains challenging due to their gigapixel resolution, strong spatial heterogeneity, and complex survival distributions. We introduce a comprehensive computational pathology framework that addresses these limitations through four complementary innovations: (1) Quantile-Gated Patch Selection for dynamically identifying prognostically relevant regions, (2) Graph-Guided Clustering to group patches by spatial and morphological similarity, (3) Hierarchical Context Attention to model both local tissue interactions and global slide-level context, and (4) an Expert-Driven Mixture of Log-Logistics module that flexibly models complex survival distributions. Across large TCGA cohorts, our method achieves state-of-the-art performance, yielding time-dependent concordance indices of 0.644 on LUAD, 0.751 on KIRC, and 0.752 on BRCA, consistently outperforming both histology-only and multimodal baselines. The framework further provides improved calibration and interpretability, advancing the use of WSIs for personalized cancer prognosis.
Mix, Align, Distil: Reliable Cross-Domain Atypical Mitosis Classification
Aug 28, 2025Abstract:Atypical mitotic figures (AMFs) are important histopathological markers yet remain challenging to identify consistently, particularly under domain shift stemming from scanner, stain, and acquisition differences. We present a simple training-time recipe for domain-robust AMF classification in MIDOG 2025 Task 2. The approach (i) increases feature diversity via style perturbations inserted at early and mid backbone stages, (ii) aligns attention-refined features across sites using weak domain labels (Scanner, Origin, Species, Tumor) through an auxiliary alignment loss, and (iii) stabilizes predictions by distilling from an exponential moving average (EMA) teacher with temperature-scaled KL divergence. On the organizer-run preliminary leaderboard for atypical mitosis classification, our submission attains balanced accuracy of 0.8762, sensitivity of 0.8873, specificity of 0.8651, and ROC AUC of 0.9499. The method incurs negligible inference-time overhead, relies only on coarse domain metadata, and delivers strong, balanced performance, positioning it as a competitive submission for the MIDOG 2025 challenge.
Federated Cross-Modal Style-Aware Prompt Generation
Aug 17, 2025Abstract:Prompt learning has propelled vision-language models like CLIP to excel in diverse tasks, making them ideal for federated learning due to computational efficiency. However, conventional approaches that rely solely on final-layer features miss out on rich multi-scale visual cues and domain-specific style variations in decentralized client data. To bridge this gap, we introduce FedCSAP (Federated Cross-Modal Style-Aware Prompt Generation). Our framework harnesses low, mid, and high-level features from CLIP's vision encoder alongside client-specific style indicators derived from batch-level statistics. By merging intricate visual details with textual context, FedCSAP produces robust, context-aware prompt tokens that are both distinct and non-redundant, thereby boosting generalization across seen and unseen classes. Operating within a federated learning paradigm, our approach ensures data privacy through local training and global aggregation, adeptly handling non-IID class distributions and diverse domain-specific styles. Comprehensive experiments on multiple image classification datasets confirm that FedCSAP outperforms existing federated prompt learning methods in both accuracy and overall generalization.
Survival Modeling from Whole Slide Images via Patch-Level Graph Clustering and Mixture Density Experts
Jul 22, 2025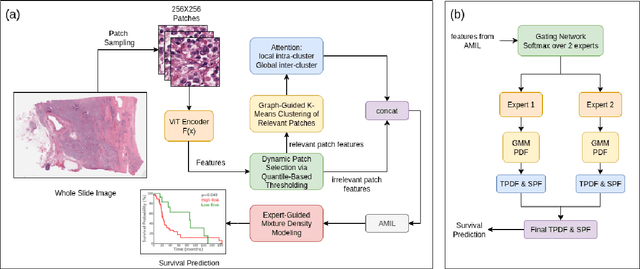
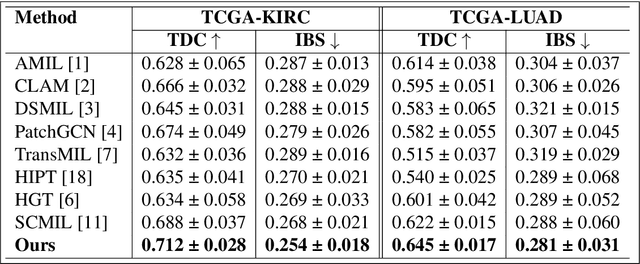
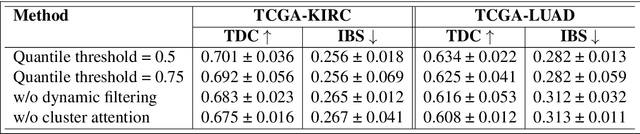
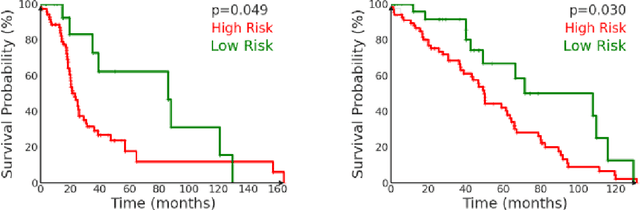
Abstract:We introduce a modular framework for predicting cancer-specific survival from whole slide pathology images (WSIs) that significantly improves upon the state-of-the-art accuracy. Our method integrating four key components. Firstly, to tackle large size of WSIs, we use dynamic patch selection via quantile-based thresholding for isolating prognostically informative tissue regions. Secondly, we use graph-guided k-means clustering to capture phenotype-level heterogeneity through spatial and morphological coherence. Thirdly, we use attention mechanisms that model both intra- and inter-cluster relationships to contextualize local features within global spatial relations between various types of tissue compartments. Finally, we use an expert-guided mixture density modeling for estimating complex survival distributions using Gaussian mixture models. The proposed model achieves a concordance index of $0.712 \pm 0.028$ and Brier score of $0.254 \pm 0.018$ on TCGA-KIRC (renal cancer), and a concordance index of $0.645 \pm 0.017$ and Brier score of $0.281 \pm 0.031$ on TCGA-LUAD (lung adenocarcinoma). These results are significantly better than the state-of-art and demonstrate predictive potential of the proposed method across diverse cancer types.
Predicting Genetic Mutations from Single-Cell Bone Marrow Images in Acute Myeloid Leukemia Using Noise-Robust Deep Learning Models
Jun 15, 2025Abstract:In this study, we propose a robust methodology for identification of myeloid blasts followed by prediction of genetic mutation in single-cell images of blasts, tackling challenges associated with label accuracy and data noise. We trained an initial binary classifier to distinguish between leukemic (blasts) and non-leukemic cells images, achieving 90 percent accuracy. To evaluate the models generalization, we applied this model to a separate large unlabeled dataset and validated the predictions with two haemato-pathologists, finding an approximate error rate of 20 percent in the leukemic and non-leukemic labels. Assuming this level of label noise, we further trained a four-class model on images predicted as blasts to classify specific mutations. The mutation labels were known for only a bag of cell images extracted from a single slide. Despite the tumor label noise, our mutation classification model achieved 85 percent accuracy across four mutation classes, demonstrating resilience to label inconsistencies. This study highlights the capability of machine learning models to work with noisy labels effectively while providing accurate, clinically relevant mutation predictions, which is promising for diagnostic applications in areas such as haemato-pathology.
A Cytology Dataset for Early Detection of Oral Squamous Cell Carcinoma
Jun 11, 2025Abstract:Oral squamous cell carcinoma OSCC is a major global health burden, particularly in several regions across Asia, Africa, and South America, where it accounts for a significant proportion of cancer cases. Early detection dramatically improves outcomes, with stage I cancers achieving up to 90 percent survival. However, traditional diagnosis based on histopathology has limited accessibility in low-resource settings because it is invasive, resource-intensive, and reliant on expert pathologists. On the other hand, oral cytology of brush biopsy offers a minimally invasive and lower cost alternative, provided that the remaining challenges, inter observer variability and unavailability of expert pathologists can be addressed using artificial intelligence. Development and validation of robust AI solutions requires access to large, labeled, and multi-source datasets to train high capacity models that generalize across domain shifts. We introduce the first large and multicenter oral cytology dataset, comprising annotated slides stained with Papanicolaou(PAP) and May-Grunwald-Giemsa(MGG) protocols, collected from ten tertiary medical centers in India. The dataset is labeled and annotated by expert pathologists for cellular anomaly classification and detection, is designed to advance AI driven diagnostic methods. By filling the gap in publicly available oral cytology datasets, this resource aims to enhance automated detection, reduce diagnostic errors, and improve early OSCC diagnosis in resource-constrained settings, ultimately contributing to reduced mortality and better patient outcomes worldwide.
FEDTAIL: Federated Long-Tailed Domain Generalization with Sharpness-Guided Gradient Matching
Jun 10, 2025Abstract:Domain Generalization (DG) seeks to train models that perform reliably on unseen target domains without access to target data during training. While recent progress in smoothing the loss landscape has improved generalization, existing methods often falter under long-tailed class distributions and conflicting optimization objectives. We introduce FedTAIL, a federated domain generalization framework that explicitly addresses these challenges through sharpness-guided, gradient-aligned optimization. Our method incorporates a gradient coherence regularizer to mitigate conflicts between classification and adversarial objectives, leading to more stable convergence. To combat class imbalance, we perform class-wise sharpness minimization and propose a curvature-aware dynamic weighting scheme that adaptively emphasizes underrepresented tail classes. Furthermore, we enhance conditional distribution alignment by integrating sharpness-aware perturbations into entropy regularization, improving robustness under domain shift. FedTAIL unifies optimization harmonization, class-aware regularization, and conditional alignment into a scalable, federated-compatible framework. Extensive evaluations across standard domain generalization benchmarks demonstrate that FedTAIL achieves state-of-the-art performance, particularly in the presence of domain shifts and label imbalance, validating its effectiveness in both centralized and federated settings. Code: https://github.com/sunnyinAI/FedTail
UniVarFL: Uniformity and Variance Regularized Federated Learning for Heterogeneous Data
Jun 09, 2025Abstract:Federated Learning (FL) often suffers from severe performance degradation when faced with non-IID data, largely due to local classifier bias. Traditional remedies such as global model regularization or layer freezing either incur high computational costs or struggle to adapt to feature shifts. In this work, we propose UniVarFL, a novel FL framework that emulates IID-like training dynamics directly at the client level, eliminating the need for global model dependency. UniVarFL leverages two complementary regularization strategies during local training: Classifier Variance Regularization, which aligns class-wise probability distributions with those expected under IID conditions, effectively mitigating local classifier bias; and Hyperspherical Uniformity Regularization, which encourages a uniform distribution of feature representations across the hypersphere, thereby enhancing the model's ability to generalize under diverse data distributions. Extensive experiments on multiple benchmark datasets demonstrate that UniVarFL outperforms existing methods in accuracy, highlighting its potential as a highly scalable and efficient solution for real-world FL deployments, especially in resource-constrained settings. Code: https://github.com/sunnyinAI/UniVarFL
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge