Garima Jain
Survival Modeling from Whole Slide Images via Patch-Level Graph Clustering and Mixture Density Experts
Jul 22, 2025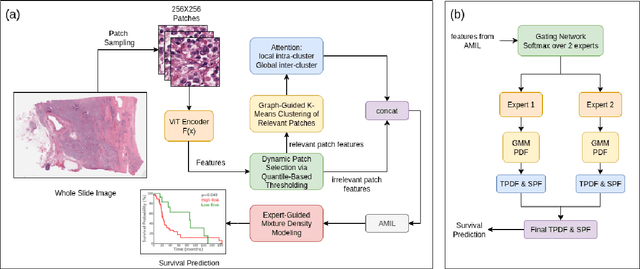
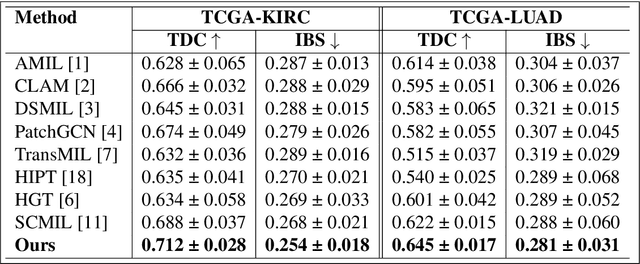
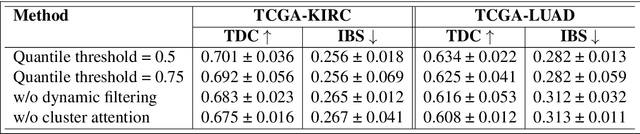
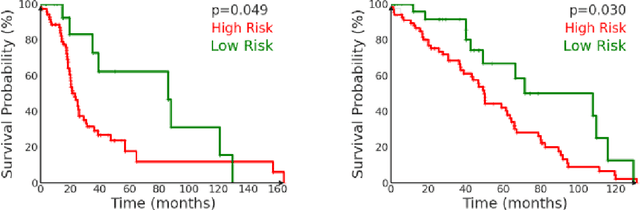
Abstract:We introduce a modular framework for predicting cancer-specific survival from whole slide pathology images (WSIs) that significantly improves upon the state-of-the-art accuracy. Our method integrating four key components. Firstly, to tackle large size of WSIs, we use dynamic patch selection via quantile-based thresholding for isolating prognostically informative tissue regions. Secondly, we use graph-guided k-means clustering to capture phenotype-level heterogeneity through spatial and morphological coherence. Thirdly, we use attention mechanisms that model both intra- and inter-cluster relationships to contextualize local features within global spatial relations between various types of tissue compartments. Finally, we use an expert-guided mixture density modeling for estimating complex survival distributions using Gaussian mixture models. The proposed model achieves a concordance index of $0.712 \pm 0.028$ and Brier score of $0.254 \pm 0.018$ on TCGA-KIRC (renal cancer), and a concordance index of $0.645 \pm 0.017$ and Brier score of $0.281 \pm 0.031$ on TCGA-LUAD (lung adenocarcinoma). These results are significantly better than the state-of-art and demonstrate predictive potential of the proposed method across diverse cancer types.
Predicting Genetic Mutations from Single-Cell Bone Marrow Images in Acute Myeloid Leukemia Using Noise-Robust Deep Learning Models
Jun 15, 2025Abstract:In this study, we propose a robust methodology for identification of myeloid blasts followed by prediction of genetic mutation in single-cell images of blasts, tackling challenges associated with label accuracy and data noise. We trained an initial binary classifier to distinguish between leukemic (blasts) and non-leukemic cells images, achieving 90 percent accuracy. To evaluate the models generalization, we applied this model to a separate large unlabeled dataset and validated the predictions with two haemato-pathologists, finding an approximate error rate of 20 percent in the leukemic and non-leukemic labels. Assuming this level of label noise, we further trained a four-class model on images predicted as blasts to classify specific mutations. The mutation labels were known for only a bag of cell images extracted from a single slide. Despite the tumor label noise, our mutation classification model achieved 85 percent accuracy across four mutation classes, demonstrating resilience to label inconsistencies. This study highlights the capability of machine learning models to work with noisy labels effectively while providing accurate, clinically relevant mutation predictions, which is promising for diagnostic applications in areas such as haemato-pathology.
A Cytology Dataset for Early Detection of Oral Squamous Cell Carcinoma
Jun 11, 2025Abstract:Oral squamous cell carcinoma OSCC is a major global health burden, particularly in several regions across Asia, Africa, and South America, where it accounts for a significant proportion of cancer cases. Early detection dramatically improves outcomes, with stage I cancers achieving up to 90 percent survival. However, traditional diagnosis based on histopathology has limited accessibility in low-resource settings because it is invasive, resource-intensive, and reliant on expert pathologists. On the other hand, oral cytology of brush biopsy offers a minimally invasive and lower cost alternative, provided that the remaining challenges, inter observer variability and unavailability of expert pathologists can be addressed using artificial intelligence. Development and validation of robust AI solutions requires access to large, labeled, and multi-source datasets to train high capacity models that generalize across domain shifts. We introduce the first large and multicenter oral cytology dataset, comprising annotated slides stained with Papanicolaou(PAP) and May-Grunwald-Giemsa(MGG) protocols, collected from ten tertiary medical centers in India. The dataset is labeled and annotated by expert pathologists for cellular anomaly classification and detection, is designed to advance AI driven diagnostic methods. By filling the gap in publicly available oral cytology datasets, this resource aims to enhance automated detection, reduce diagnostic errors, and improve early OSCC diagnosis in resource-constrained settings, ultimately contributing to reduced mortality and better patient outcomes worldwide.
Foundation Models for Time Series: A Survey
Apr 05, 2025Abstract:Transformer-based foundation models have emerged as a dominant paradigm in time series analysis, offering unprecedented capabilities in tasks such as forecasting, anomaly detection, classification, trend analysis and many more time series analytical tasks. This survey provides a comprehensive overview of the current state of the art pre-trained foundation models, introducing a novel taxonomy to categorize them across several dimensions. Specifically, we classify models by their architecture design, distinguishing between those leveraging patch-based representations and those operating directly on raw sequences. The taxonomy further includes whether the models provide probabilistic or deterministic predictions, and whether they are designed to work with univariate time series or can handle multivariate time series out of the box. Additionally, the taxonomy encompasses model scale and complexity, highlighting differences between lightweight architectures and large-scale foundation models. A unique aspect of this survey is its categorization by the type of objective function employed during training phase. By synthesizing these perspectives, this survey serves as a resource for researchers and practitioners, providing insights into current trends and identifying promising directions for future research in transformer-based time series modeling.
Classification and Morphological Analysis of DLBCL Subtypes in H\&E-Stained Slides
Nov 13, 2024
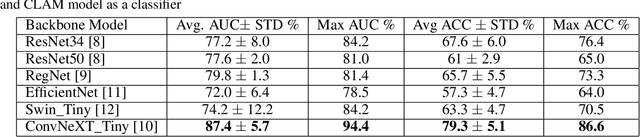
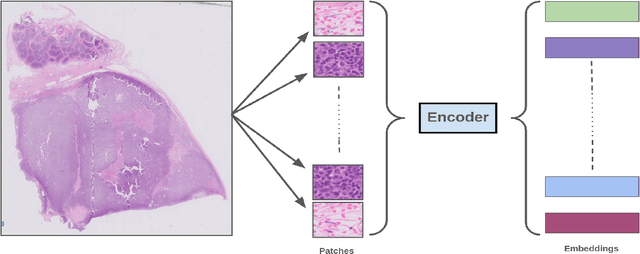
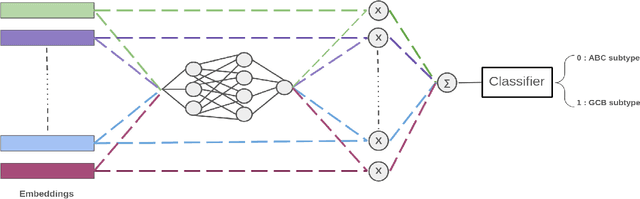
Abstract:We address the challenge of automated classification of diffuse large B-cell lymphoma (DLBCL) into its two primary subtypes: activated B-cell-like (ABC) and germinal center B-cell-like (GCB). Accurate classification between these subtypes is essential for determining the appropriate therapeutic strategy, given their distinct molecular profiles and treatment responses. Our proposed deep learning model demonstrates robust performance, achieving an average area under the curve (AUC) of (87.4 pm 5.7)\% during cross-validation. It shows a high positive predictive value (PPV), highlighting its potential for clinical application, such as triaging for molecular testing. To gain biological insights, we performed an analysis of morphological features of ABC and GCB subtypes. We segmented cell nuclei using a pre-trained deep neural network and compared the statistics of geometric and color features for ABC and GCB. We found that the distributions of these features were not very different for the two subtypes, which suggests that the visual differences between them are more subtle. These results underscore the potential of our method to assist in more precise subtype classification and can contribute to improved treatment management and outcomes for patients of DLBCL.
Semantic Segmentation Based Quality Control of Histopathology Whole Slide Images
Oct 04, 2024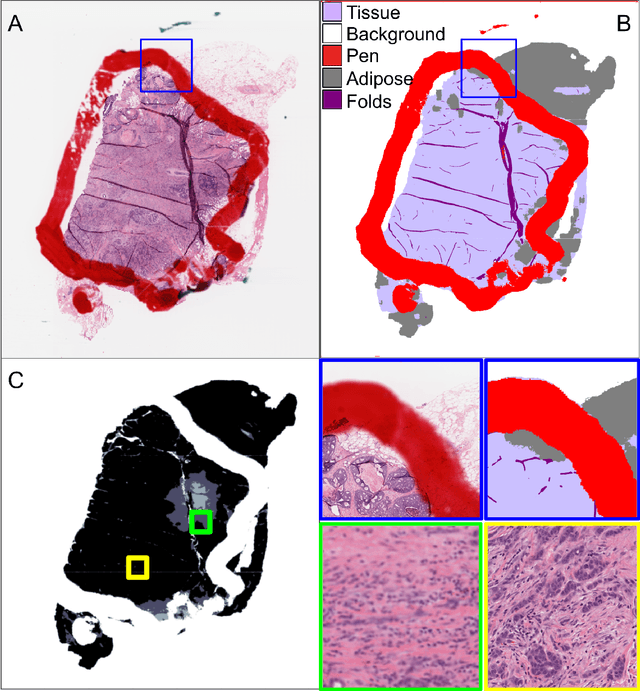
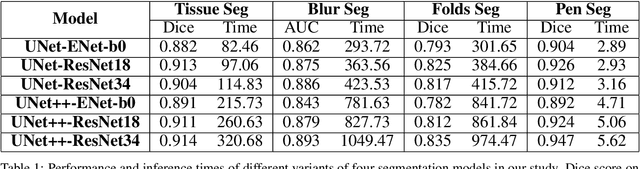
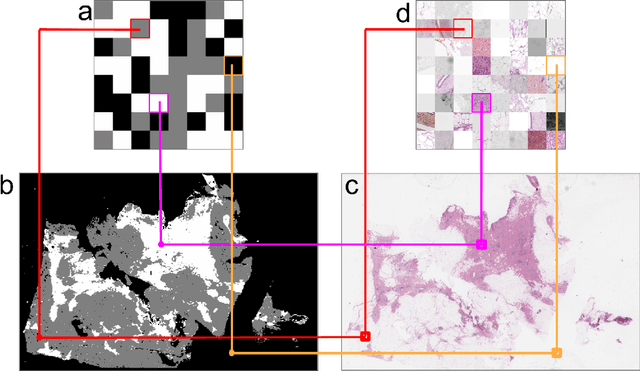
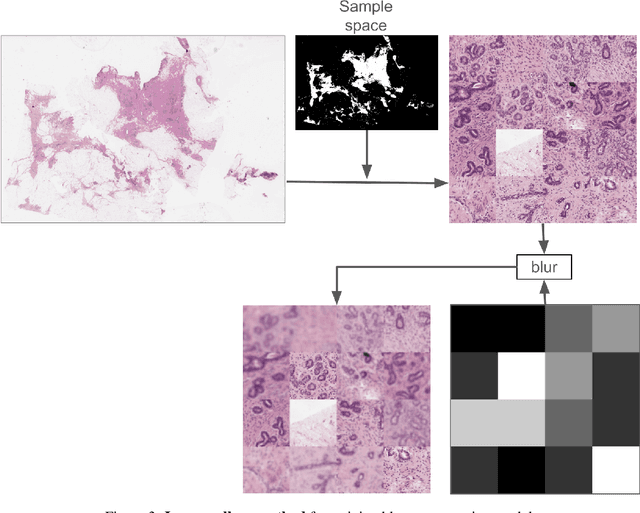
Abstract:We developed a software pipeline for quality control (QC) of histopathology whole slide images (WSIs) that segments various regions, such as blurs of different levels, tissue regions, tissue folds, and pen marks. Given the necessity and increasing availability of GPUs for processing WSIs, the proposed pipeline comprises multiple lightweight deep learning models to strike a balance between accuracy and speed. The pipeline was evaluated in all TCGAs, which is the largest publicly available WSI dataset containing more than 11,000 histopathological images from 28 organs. It was compared to a previous work, which was not based on deep learning, and it showed consistent improvement in segmentation results across organs. To minimize annotation effort for tissue and blur segmentation, annotated images were automatically prepared by mosaicking patches (sub-images) from various WSIs whose labels were identified using a patch classification tool HistoROI. Due to the generality of our trained QC pipeline and its extensive testing the potential impact of this work is broad. It can be used for automated pre-processing any WSI cohort to enhance the accuracy and reliability of large-scale histopathology image analysis for both research and clinical use. We have made the trained models, training scripts, training data, and inference results publicly available at https://github.com/abhijeetptl5/wsisegqc, which should enable the research community to use the pipeline right out of the box or further customize it to new datasets and applications in the future.
HER2 and FISH Status Prediction in Breast Biopsy H&E-Stained Images Using Deep Learning
Aug 25, 2024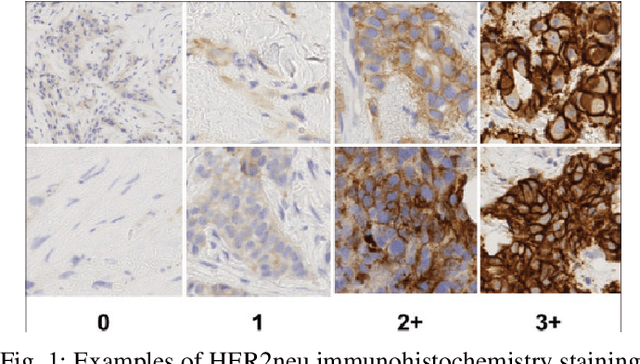
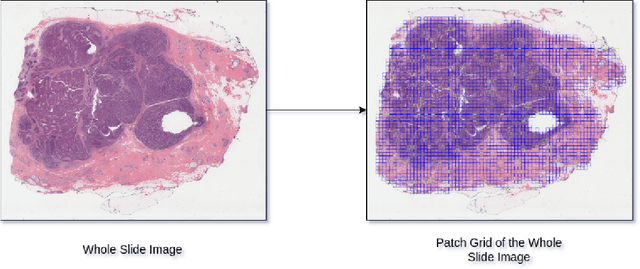

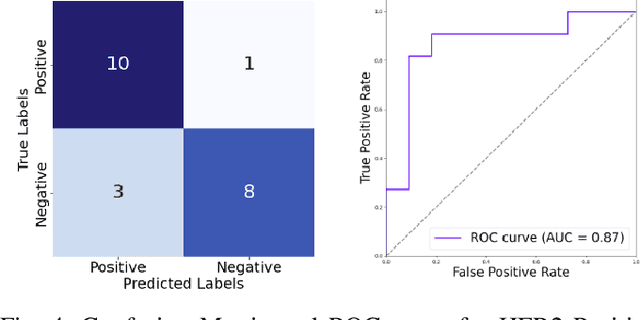
Abstract:The current standard for detecting human epidermal growth factor receptor 2 (HER2) status in breast cancer patients relies on HER2 amplification, identified through fluorescence in situ hybridization (FISH) or immunohistochemistry (IHC). However, hematoxylin and eosin (H\&E) tumor stains are more widely available, and accurately predicting HER2 status using H\&E could reduce costs and expedite treatment selection. Deep Learning algorithms for H&E have shown effectiveness in predicting various cancer features and clinical outcomes, including moderate success in HER2 status prediction. In this work, we employed a customized weak supervision classification technique combined with MoCo-v2 contrastive learning to predict HER2 status. We trained our pipeline on 182 publicly available H&E Whole Slide Images (WSIs) from The Cancer Genome Atlas (TCGA), for which annotations by the pathology team at Yale School of Medicine are publicly available. Our pipeline achieved an Area Under the Curve (AUC) of 0.85 across four different test folds. Additionally, we tested our model on 44 H&E slides from the TCGA-BRCA dataset, which had an HER2 score of 2+ and included corresponding HER2 status and FISH test results. These cases are considered equivocal for IHC, requiring an expensive FISH test on their IHC slides for disambiguation. Our pipeline demonstrated an AUC of 0.81 on these challenging H&E slides. Reducing the need for FISH test can have significant implications in cancer treatment equity for underserved populations.
Bioimpedance a Diagnostic Tool for Tobacco Induced Oral Lesions: a Mixed Model cross-sectional study
Aug 21, 2024Abstract:Introduction: Electrical impedance spectroscopy (EIS) has recently developed as a novel diagnostic device for screening and evaluating cervical dysplasia, prostate cancer, breast cancer and basal cell carcinoma. The current study aimed to validate and evaluate bioimpedance as a diagnostic tool for tobacco-induced oral lesions. Methodology: The study comprised 50 OSCC and OPMD tissue specimens for in-vitro study and 320 subjects for in vivo study. Bioimpedance device prepared and calibrated. EIS measurements were done for the habit and control groups and were compared. Results: The impedance value in the control group was significantly higher compared to the OPMD and OSCC groups. Diagnosis based on BIS measurements has a sensitivity of 95.9% and a specificity of 86.7%. Conclusion: Bioimpedance device can help in decision-making for differentiating OPMD and OSCC cases and their management, especially in primary healthcare settings. Keywords: Impedance, Cancer, Diagnosis, Device, Community
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge