Semantic Segmentation Based Quality Control of Histopathology Whole Slide Images
Paper and Code
Oct 04, 2024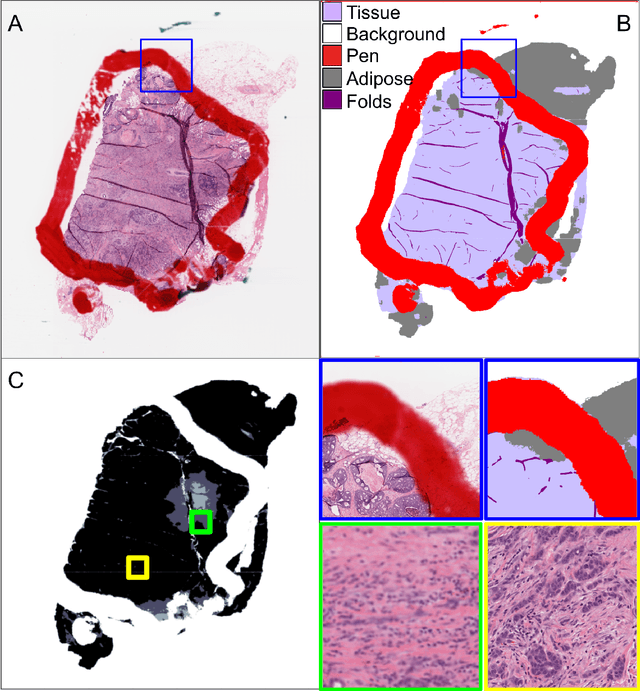
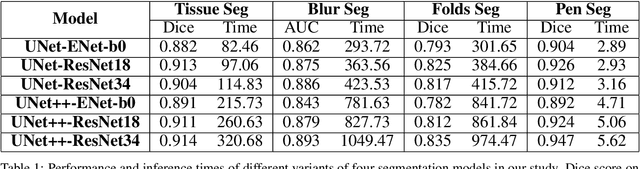
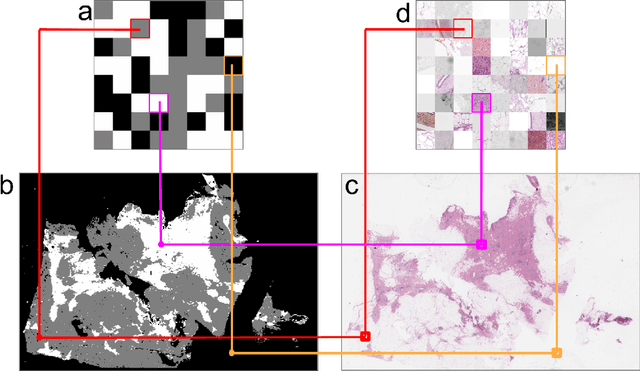
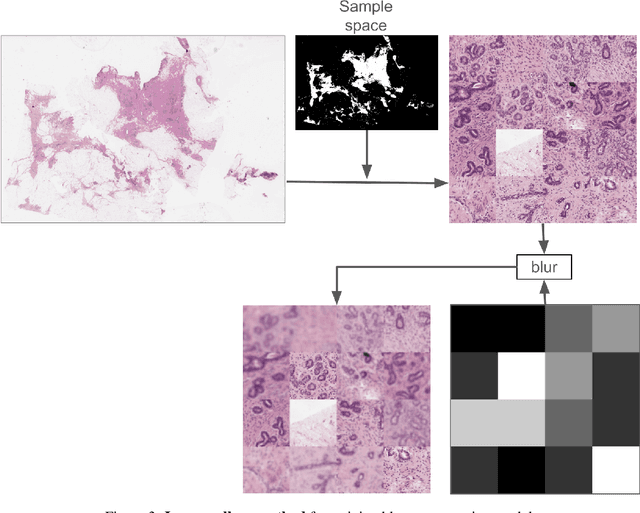
We developed a software pipeline for quality control (QC) of histopathology whole slide images (WSIs) that segments various regions, such as blurs of different levels, tissue regions, tissue folds, and pen marks. Given the necessity and increasing availability of GPUs for processing WSIs, the proposed pipeline comprises multiple lightweight deep learning models to strike a balance between accuracy and speed. The pipeline was evaluated in all TCGAs, which is the largest publicly available WSI dataset containing more than 11,000 histopathological images from 28 organs. It was compared to a previous work, which was not based on deep learning, and it showed consistent improvement in segmentation results across organs. To minimize annotation effort for tissue and blur segmentation, annotated images were automatically prepared by mosaicking patches (sub-images) from various WSIs whose labels were identified using a patch classification tool HistoROI. Due to the generality of our trained QC pipeline and its extensive testing the potential impact of this work is broad. It can be used for automated pre-processing any WSI cohort to enhance the accuracy and reliability of large-scale histopathology image analysis for both research and clinical use. We have made the trained models, training scripts, training data, and inference results publicly available at https://github.com/abhijeetptl5/wsisegqc, which should enable the research community to use the pipeline right out of the box or further customize it to new datasets and applications in the future.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge