Subhash Yadav
IMPROVE: Improving Medical Plausibility without Reliance on HumanValidation -- An Enhanced Prototype-Guided Diffusion Framework
Nov 26, 2024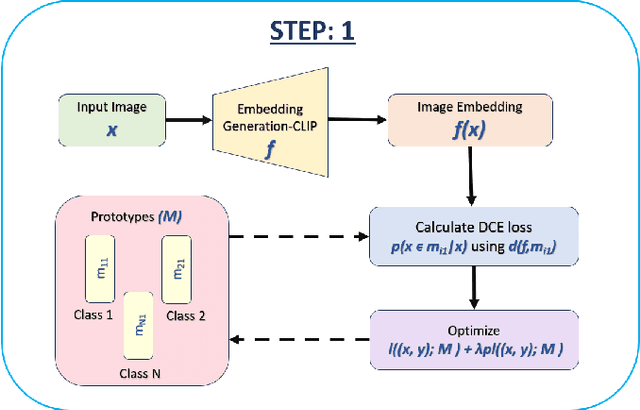

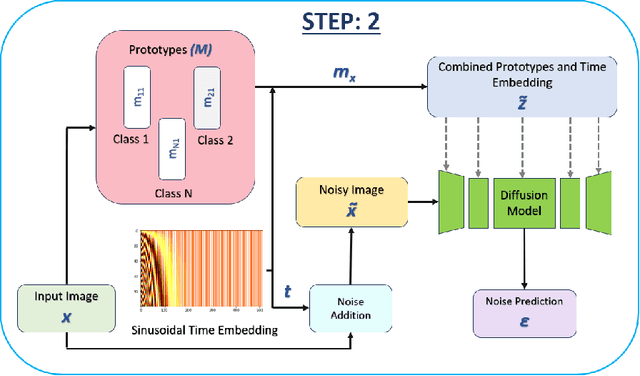
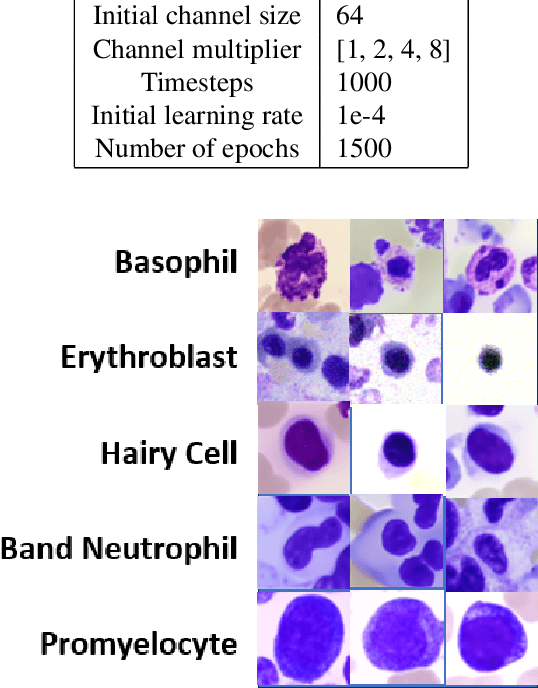
Abstract:Generative models have proven to be very effective in generating synthetic medical images and find applications in downstream tasks such as enhancing rare disease datasets, long-tailed dataset augmentation, and scaling machine learning algorithms. For medical applications, the synthetically generated medical images by such models are still reasonable in quality when evaluated based on traditional metrics such as FID score, precision, and recall. However, these metrics fail to capture the medical/biological plausibility of the generated images. Human expert feedback has been used to get biological plausibility which demonstrates that these generated images have very low plausibility. Recently, the research community has further integrated this human feedback through Reinforcement Learning from Human Feedback(RLHF), which generates more medically plausible images. However, incorporating human feedback is a costly and slow process. In this work, we propose a novel approach to improve the medical plausibility of generated images without the need for human feedback. We introduce IMPROVE:Improving Medical Plausibility without Reliance on Human Validation - An Enhanced Prototype-Guided Diffusion Framework, a prototype-guided diffusion process for medical image generation and show that it substantially enhances the biological plausibility of the generated medical images without the need for any human feedback. We perform experiments on Bone Marrow and HAM10000 datasets and show that medical accuracy can be substantially increased without human feedback.
Classification and Morphological Analysis of DLBCL Subtypes in H\&E-Stained Slides
Nov 13, 2024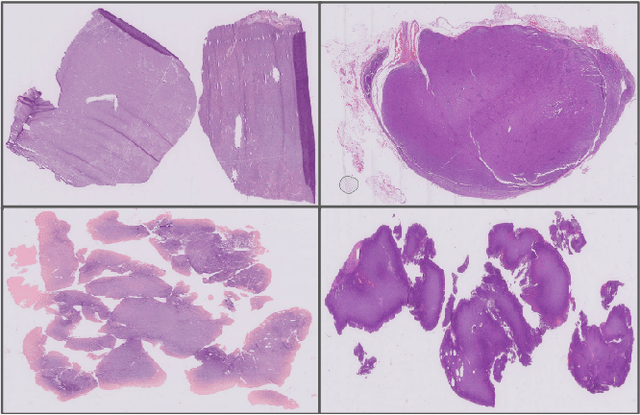
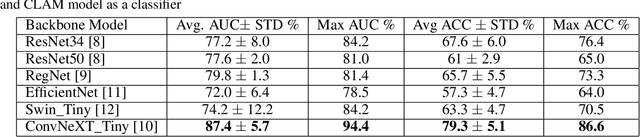
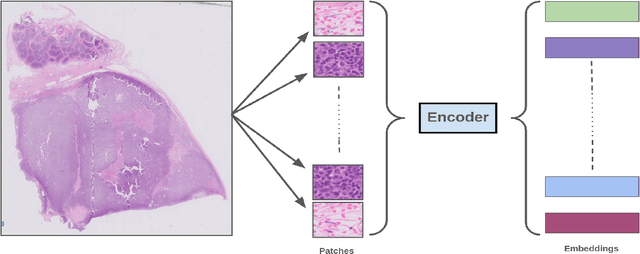
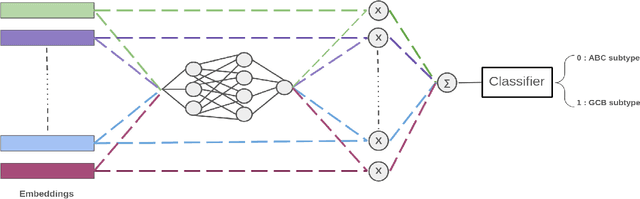
Abstract:We address the challenge of automated classification of diffuse large B-cell lymphoma (DLBCL) into its two primary subtypes: activated B-cell-like (ABC) and germinal center B-cell-like (GCB). Accurate classification between these subtypes is essential for determining the appropriate therapeutic strategy, given their distinct molecular profiles and treatment responses. Our proposed deep learning model demonstrates robust performance, achieving an average area under the curve (AUC) of (87.4 pm 5.7)\% during cross-validation. It shows a high positive predictive value (PPV), highlighting its potential for clinical application, such as triaging for molecular testing. To gain biological insights, we performed an analysis of morphological features of ABC and GCB subtypes. We segmented cell nuclei using a pre-trained deep neural network and compared the statistics of geometric and color features for ABC and GCB. We found that the distributions of these features were not very different for the two subtypes, which suggests that the visual differences between them are more subtle. These results underscore the potential of our method to assist in more precise subtype classification and can contribute to improved treatment management and outcomes for patients of DLBCL.
Efficient Quality Control of Whole Slide Pathology Images with Human-in-the-loop Training
Sep 29, 2024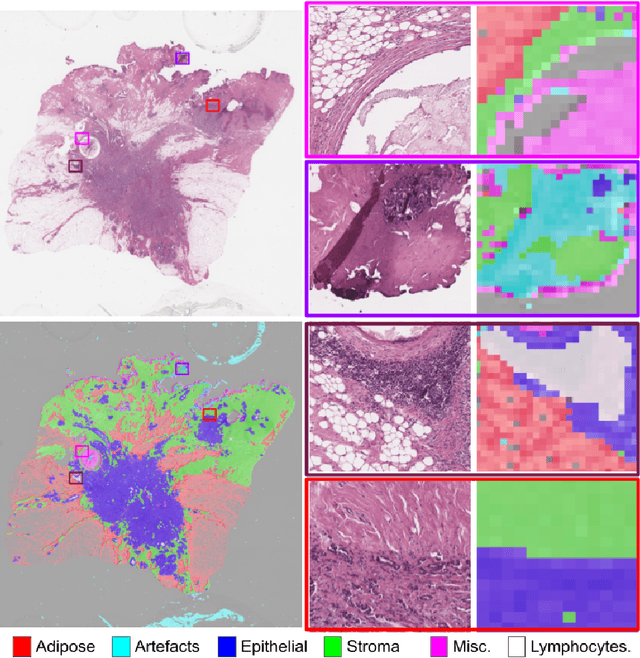
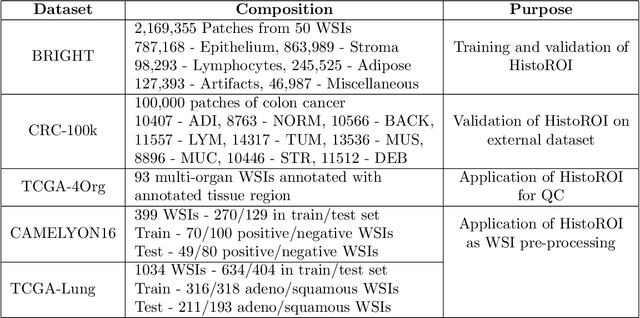
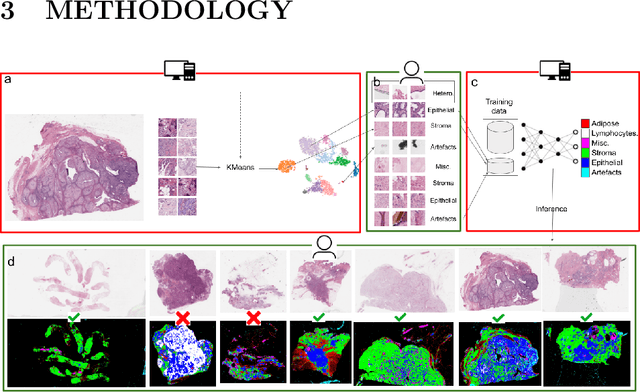
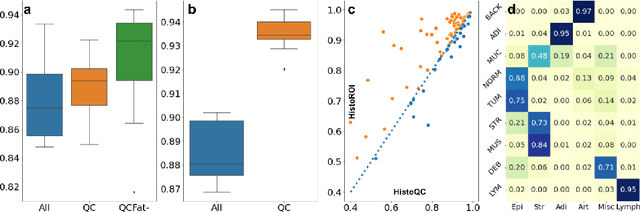
Abstract:Histopathology whole slide images (WSIs) are being widely used to develop deep learning-based diagnostic solutions, especially for precision oncology. Most of these diagnostic softwares are vulnerable to biases and impurities in the training and test data which can lead to inaccurate diagnoses. For instance, WSIs contain multiple types of tissue regions, at least some of which might not be relevant to the diagnosis. We introduce HistoROI, a robust yet lightweight deep learning-based classifier to segregate WSI into six broad tissue regions -- epithelium, stroma, lymphocytes, adipose, artifacts, and miscellaneous. HistoROI is trained using a novel human-in-the-loop and active learning paradigm that ensures variations in training data for labeling-efficient generalization. HistoROI consistently performs well across multiple organs, despite being trained on only a single dataset, demonstrating strong generalization. Further, we have examined the utility of HistoROI in improving the performance of downstream deep learning-based tasks using the CAMELYON breast cancer lymph node and TCGA lung cancer datasets. For the former dataset, the area under the receiver operating characteristic curve (AUC) for metastasis versus normal tissue of a neural network trained using weakly supervised learning increased from 0.88 to 0.92 by filtering the data using HistoROI. Similarly, the AUC increased from 0.88 to 0.93 for the classification between adenocarcinoma and squamous cell carcinoma on the lung cancer dataset. We also found that the performance of the HistoROI improves upon HistoQC for artifact detection on a test dataset of 93 annotated WSIs. The limitations of the proposed model are analyzed, and potential extensions are also discussed.
* 18 pages
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge