Kshitij Jadhav
RadJEPA: Radiology Encoder for Chest X-Rays via Joint Embedding Predictive Architecture
Jan 22, 2026Abstract:Recent advances in medical vision language models guide the learning of visual representations; however, this form of supervision is constrained by the availability of paired image text data, raising the question of whether robust radiology encoders can be learned without relying on language supervision. In this work, we introduce RadJEPA, a self-supervised framework built on a Joint Embedding Predictive Architecture that learns without language supervision. Pre-trained solely on unlabeled chest X-ray images, the model learns to predict latent representations of masked image regions. This predictive objective differs fundamentally from both image text pre-training and DINO-style self-distillation: rather than aligning global representations across views or modalities, RadJEPA explicitly models latent-space prediction. We evaluate the learned encoder on disease classification, semantic segmentation, and report generation tasks. Across benchmarks, RadJEPA achieves performance exceeding state-of-the-art approaches, including Rad-DINO.
Broken Words, Broken Performance: Effect of Tokenization on Performance of LLMs
Dec 26, 2025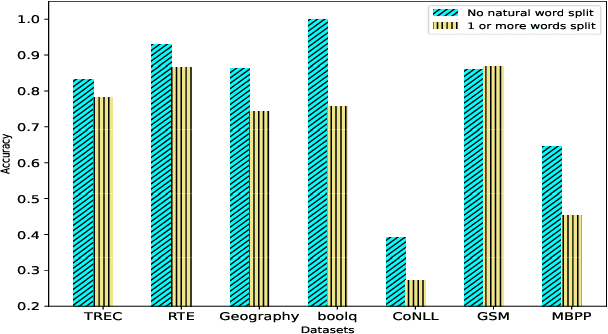
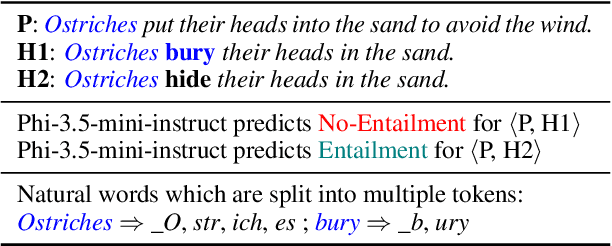

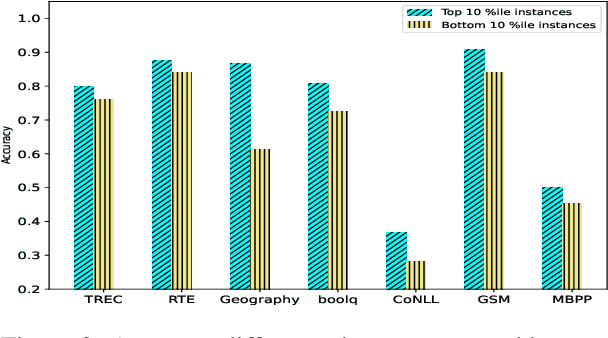
Abstract:Tokenization is the first step in training any Large Language Model (LLM), where the text is split into a sequence of tokens as per the model's fixed vocabulary. This tokenization in LLMs is different from the traditional tokenization in NLP where the text is split into a sequence of "natural" words. In LLMs, a natural word may also be broken into multiple tokens due to limited vocabulary size of the LLMs (e.g., Mistral's tokenizer splits "martial" into "mart" and "ial"). In this paper, we hypothesize that such breaking of natural words negatively impacts LLM performance on various NLP tasks. To quantify this effect, we propose a set of penalty functions that compute a tokenization penalty for a given text for a specific LLM, indicating how "bad" the tokenization is. We establish statistical significance of our hypothesis on multiple NLP tasks for a set of different LLMs.
Interpretable Few-Shot Retinal Disease Diagnosis with Concept-Guided Prompting of Vision-Language Models
Mar 04, 2025Abstract:Recent advancements in deep learning have shown significant potential for classifying retinal diseases using color fundus images. However, existing works predominantly rely exclusively on image data, lack interpretability in their diagnostic decisions, and treat medical professionals primarily as annotators for ground truth labeling. To fill this gap, we implement two key strategies: extracting interpretable concepts of retinal diseases using the knowledge base of GPT models and incorporating these concepts as a language component in prompt-learning to train vision-language (VL) models with both fundus images and their associated concepts. Our method not only improves retinal disease classification but also enriches few-shot and zero-shot detection (novel disease detection), while offering the added benefit of concept-based model interpretability. Our extensive evaluation across two diverse retinal fundus image datasets illustrates substantial performance gains in VL-model based few-shot methodologies through our concept integration approach, demonstrating an average improvement of approximately 5.8\% and 2.7\% mean average precision for 16-shot learning and zero-shot (novel class) detection respectively. Our method marks a pivotal step towards interpretable and efficient retinal disease recognition for real-world clinical applications.
IMPROVE: Improving Medical Plausibility without Reliance on HumanValidation -- An Enhanced Prototype-Guided Diffusion Framework
Nov 26, 2024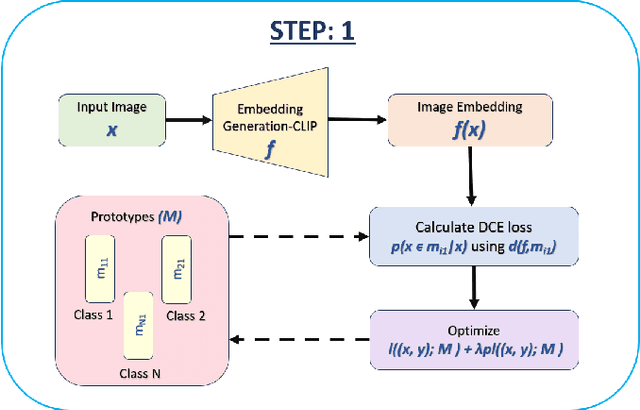

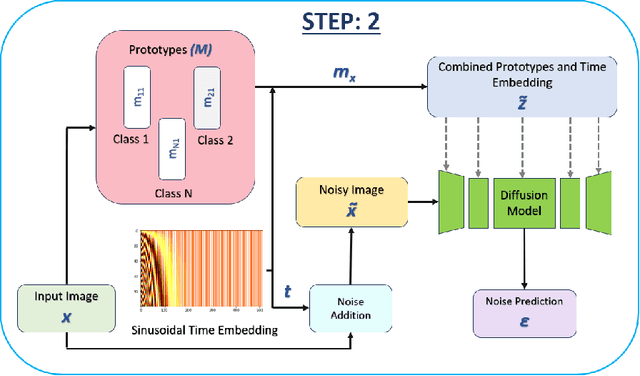
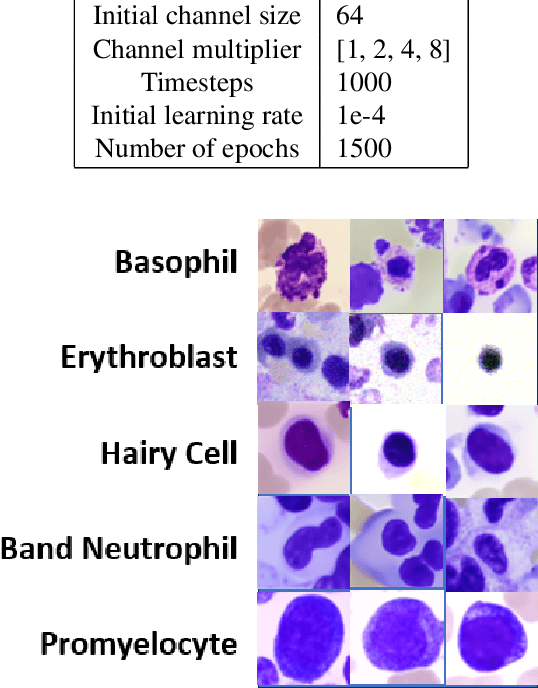
Abstract:Generative models have proven to be very effective in generating synthetic medical images and find applications in downstream tasks such as enhancing rare disease datasets, long-tailed dataset augmentation, and scaling machine learning algorithms. For medical applications, the synthetically generated medical images by such models are still reasonable in quality when evaluated based on traditional metrics such as FID score, precision, and recall. However, these metrics fail to capture the medical/biological plausibility of the generated images. Human expert feedback has been used to get biological plausibility which demonstrates that these generated images have very low plausibility. Recently, the research community has further integrated this human feedback through Reinforcement Learning from Human Feedback(RLHF), which generates more medically plausible images. However, incorporating human feedback is a costly and slow process. In this work, we propose a novel approach to improve the medical plausibility of generated images without the need for human feedback. We introduce IMPROVE:Improving Medical Plausibility without Reliance on Human Validation - An Enhanced Prototype-Guided Diffusion Framework, a prototype-guided diffusion process for medical image generation and show that it substantially enhances the biological plausibility of the generated medical images without the need for any human feedback. We perform experiments on Bone Marrow and HAM10000 datasets and show that medical accuracy can be substantially increased without human feedback.
PRISM: Privacy-preserving Inter-Site MRI Harmonization via Disentangled Representation Learning
Nov 10, 2024Abstract:Multi-site MRI studies often suffer from site-specific variations arising from differences in methodology, hardware, and acquisition protocols, thereby compromising accuracy and reliability in clinical AI/ML tasks. We present PRISM (Privacy-preserving Inter-Site MRI Harmonization), a novel Deep Learning framework for harmonizing structural brain MRI across multiple sites while preserving data privacy. PRISM employs a dual-branch autoencoder with contrastive learning and variational inference to disentangle anatomical features from style and site-specific variations, enabling unpaired image translation without traveling subjects or multiple MRI modalities. Our modular design allows harmonization to any target site and seamless integration of new sites without the need for retraining or fine-tuning. Using multi-site structural MRI data, we demonstrate PRISM's effectiveness in downstream tasks such as brain tissue segmentation and validate its harmonization performance through multiple experiments. Our framework addresses key challenges in medical AI/ML, including data privacy, distribution shifts, model generalizability and interpretability. Code is available at https://github.com/saranggalada/PRISM
MIRAGE: Multimodal Identification and Recognition of Annotations in Indian General Prescriptions
Oct 13, 2024Abstract:Hospitals generate thousands of handwritten prescriptions, a practice that remains prevalent despite the availability of Electronic Medical Records (EMR). This method of record-keeping hinders the examination of long-term medication effects, impedes statistical analysis, and makes the retrieval of records challenging. Handwritten prescriptions pose a unique challenge, requiring specialized data for training models to recognize medications and their patterns of recommendation. While current handwriting recognition approaches typically employ 2-D LSTMs, recent studies have explored the use of Large Language Models (LLMs) for Optical Character Recognition (OCR). Building on this approach, we focus on extracting medication names from medical records. Our methodology MIRAGE (Multimodal Identification and Recognition of Annotations in indian GEneral prescriptions) involves fine-tuning the LLaVA 1.6 and Idefics2 models. Our research utilizes a dataset provided by Medyug Technology, consisting of 743,118 fully annotated high-resolution simulated medical records from 1,133 doctors across India. We demonstrate that our methodology exhibits 82% accuracy in medication name and dosage extraction. We provide a detailed account of our research methodology and results, notes about HWR with Multimodal LLMs, and release a small dataset of 100 medical records with labels.
One Shot GANs for Long Tail Problem in Skin Lesion Dataset using novel content space assessment metric
Sep 30, 2024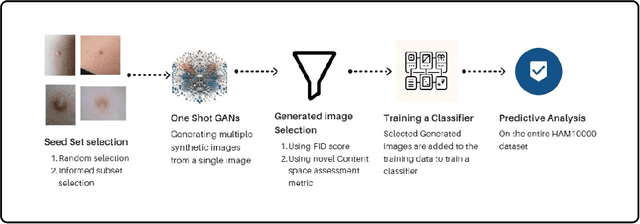
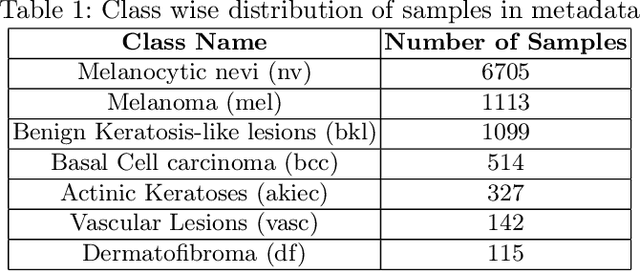
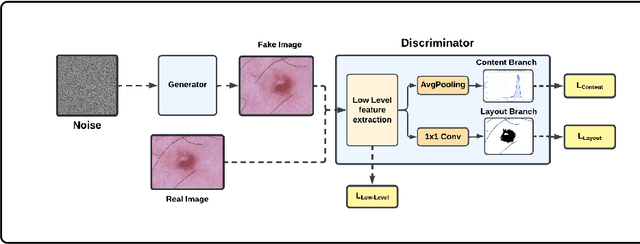
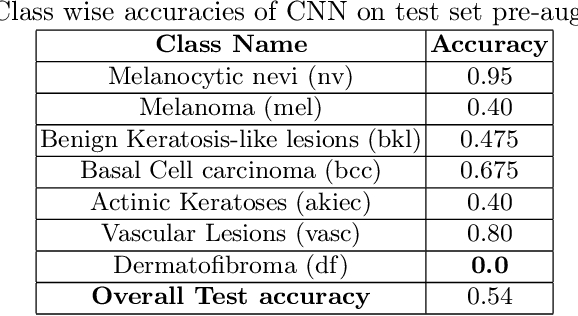
Abstract:Long tail problems frequently arise in the medical field, particularly due to the scarcity of medical data for rare conditions. This scarcity often leads to models overfitting on such limited samples. Consequently, when training models on datasets with heavily skewed classes, where the number of samples varies significantly, a problem emerges. Training on such imbalanced datasets can result in selective detection, where a model accurately identifies images belonging to the majority classes but disregards those from minority classes. This causes the model to lack generalizability, preventing its use on newer data. This poses a significant challenge in developing image detection and diagnosis models for medical image datasets. To address this challenge, the One Shot GANs model was employed to augment the tail class of HAM10000 dataset by generating additional samples. Furthermore, to enhance accuracy, a novel metric tailored to suit One Shot GANs was utilized.
Harnessing Shared Relations via Multimodal Mixup Contrastive Learning for Multimodal Classification
Sep 26, 2024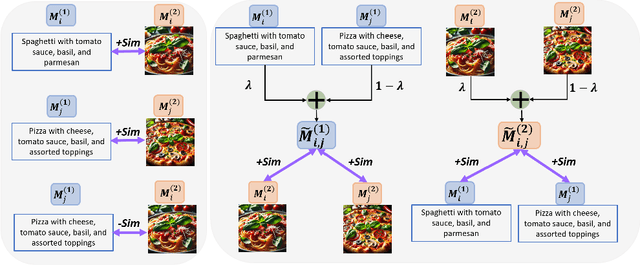
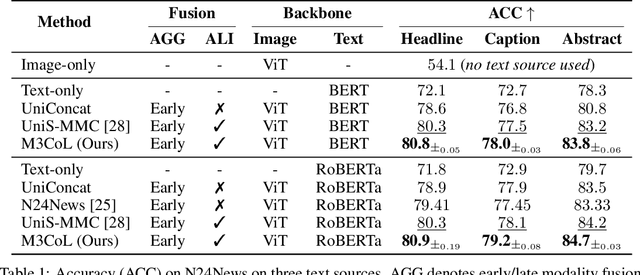
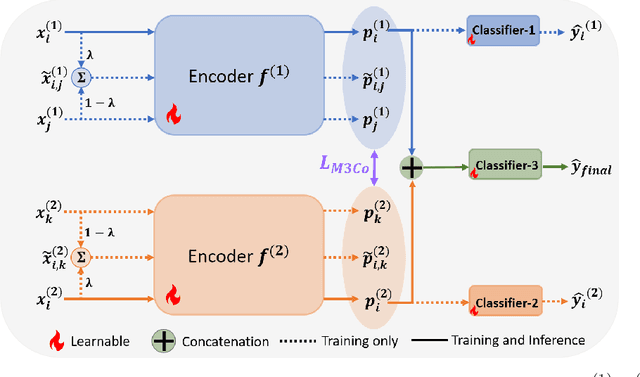
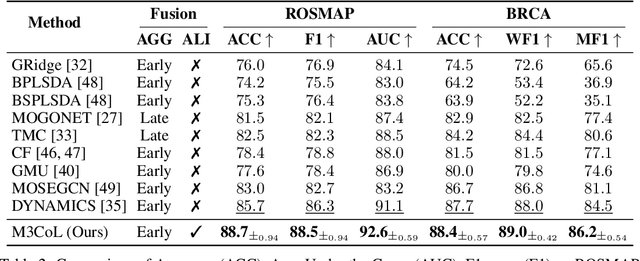
Abstract:Deep multimodal learning has shown remarkable success by leveraging contrastive learning to capture explicit one-to-one relations across modalities. However, real-world data often exhibits shared relations beyond simple pairwise associations. We propose M3CoL, a Multimodal Mixup Contrastive Learning approach to capture nuanced shared relations inherent in multimodal data. Our key contribution is a Mixup-based contrastive loss that learns robust representations by aligning mixed samples from one modality with their corresponding samples from other modalities thereby capturing shared relations between them. For multimodal classification tasks, we introduce a framework that integrates a fusion module with unimodal prediction modules for auxiliary supervision during training, complemented by our proposed Mixup-based contrastive loss. Through extensive experiments on diverse datasets (N24News, ROSMAP, BRCA, and Food-101), we demonstrate that M3CoL effectively captures shared multimodal relations and generalizes across domains. It outperforms state-of-the-art methods on N24News, ROSMAP, and BRCA, while achieving comparable performance on Food-101. Our work highlights the significance of learning shared relations for robust multimodal learning, opening up promising avenues for future research.
MedPromptExtract (Medical Data Extraction Tool): Anonymization and Hi-fidelity Automated data extraction using NLP and prompt engineering
May 04, 2024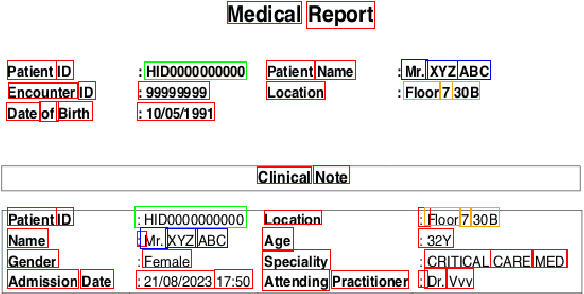

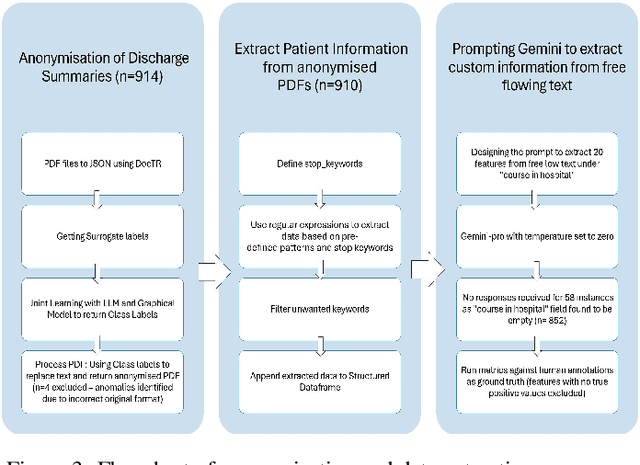
Abstract:A major roadblock in the seamless digitization of medical records remains the lack of interoperability of existing records. Extracting relevant medical information required for further treatment planning or even research is a time consuming labour intensive task involving the much valuable time of doctors. In this demo paper we present, MedPromptExtract an automated tool using a combination of semi supervised learning, large language models, natural lanuguage processing and prompt engineering to convert unstructured medical records to structured data which is amenable to further analysis.
INSITE: labelling medical images using submodular functions and semi-supervised data programming
Feb 11, 2024
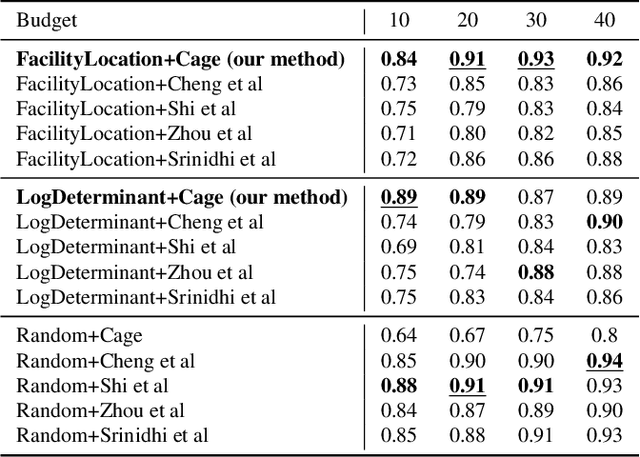
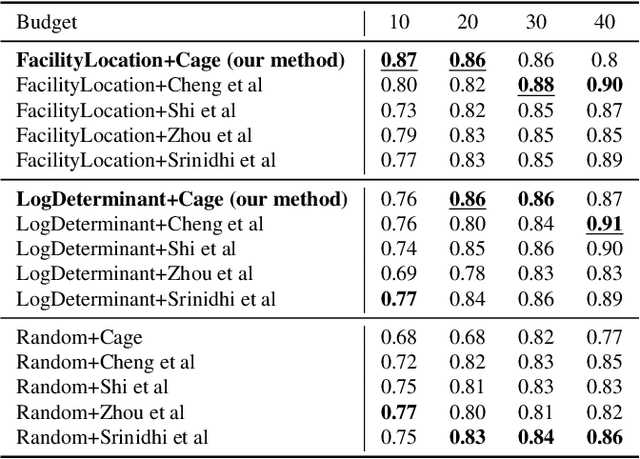
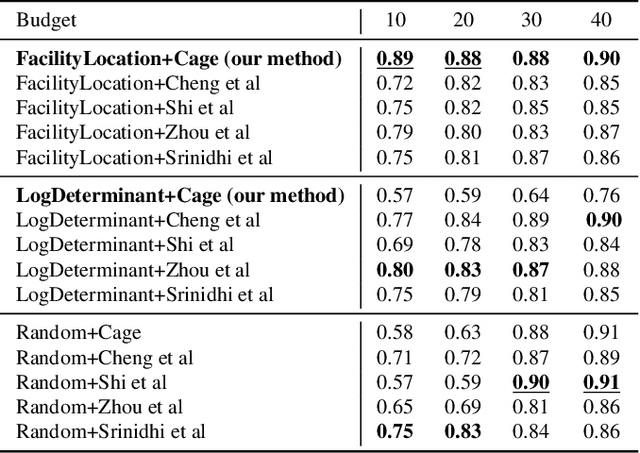
Abstract:The necessity of large amounts of labeled data to train deep models, especially in medical imaging creates an implementation bottleneck in resource-constrained settings. In Insite (labelINg medical imageS usIng submodular funcTions and sEmi-supervised data programming) we apply informed subset selection to identify a small number of most representative or diverse images from a huge pool of unlabelled data subsequently annotated by a domain expert. The newly annotated images are then used as exemplars to develop several data programming-driven labeling functions. These labelling functions output a predicted-label and a similarity score when given an unlabelled image as an input. A consensus is brought amongst the outputs of these labeling functions by using a label aggregator function to assign the final predicted label to each unlabelled data point. We demonstrate that informed subset selection followed by semi-supervised data programming methods using these images as exemplars perform better than other state-of-the-art semi-supervised methods. Further, for the first time we demonstrate that this can be achieved through a small set of images used as exemplars.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge