Raghav Singhal
Michael Pokorny
Apertus: Democratizing Open and Compliant LLMs for Global Language Environments
Sep 17, 2025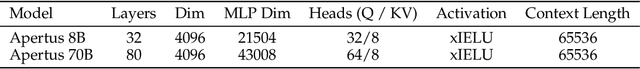
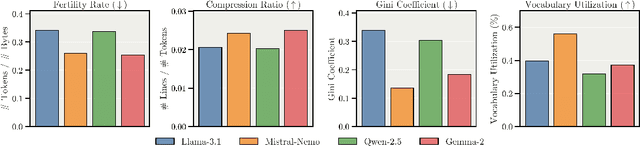
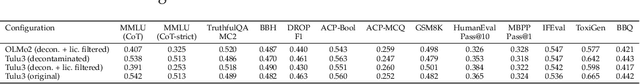
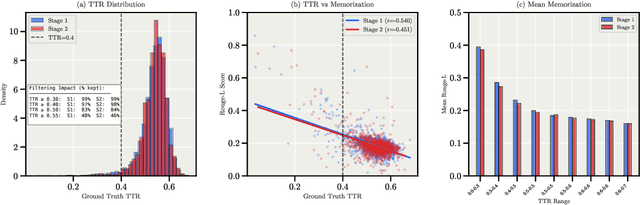
Abstract:We present Apertus, a fully open suite of large language models (LLMs) designed to address two systemic shortcomings in today's open model ecosystem: data compliance and multilingual representation. Unlike many prior models that release weights without reproducible data pipelines or regard for content-owner rights, Apertus models are pretrained exclusively on openly available data, retroactively respecting robots.txt exclusions and filtering for non-permissive, toxic, and personally identifiable content. To mitigate risks of memorization, we adopt the Goldfish objective during pretraining, strongly suppressing verbatim recall of data while retaining downstream task performance. The Apertus models also expand multilingual coverage, training on 15T tokens from over 1800 languages, with ~40% of pretraining data allocated to non-English content. Released at 8B and 70B scales, Apertus approaches state-of-the-art results among fully open models on multilingual benchmarks, rivalling or surpassing open-weight counterparts. Beyond model weights, we release all scientific artifacts from our development cycle with a permissive license, including data preparation scripts, checkpoints, evaluation suites, and training code, enabling transparent audit and extension.
Safety Subspaces are Not Distinct: A Fine-Tuning Case Study
May 20, 2025Abstract:Large Language Models (LLMs) rely on safety alignment to produce socially acceptable responses. This is typically achieved through instruction tuning and reinforcement learning from human feedback. However, this alignment is known to be brittle: further fine-tuning, even on benign or lightly contaminated data, can degrade safety and reintroduce harmful behaviors. A growing body of work suggests that alignment may correspond to identifiable geometric directions in weight space, forming subspaces that could, in principle, be isolated or preserved to defend against misalignment. In this work, we conduct a comprehensive empirical study of this geometric perspective. We examine whether safety-relevant behavior is concentrated in specific subspaces, whether it can be separated from general-purpose learning, and whether harmfulness arises from distinguishable patterns in internal representations. Across both parameter and activation space, our findings are consistent: subspaces that amplify safe behaviors also amplify unsafe ones, and prompts with different safety implications activate overlapping representations. We find no evidence of a subspace that selectively governs safety. These results challenge the assumption that alignment is geometrically localized. Rather than residing in distinct directions, safety appears to emerge from entangled, high-impact components of the model's broader learning dynamics. This suggests that subspace-based defenses may face fundamental limitations and underscores the need for alternative strategies to preserve alignment under continued training. We corroborate these findings through multiple experiments on five open-source LLMs. Our code is publicly available at: https://github.com/CERT-Lab/safety-subspaces.
ABBA: Highly Expressive Hadamard Product Adaptation for Large Language Models
May 20, 2025Abstract:Large Language Models have demonstrated strong performance across a wide range of tasks, but adapting them efficiently to new domains remains a key challenge. Parameter-Efficient Fine-Tuning (PEFT) methods address this by introducing lightweight, trainable modules while keeping most pre-trained weights fixed. The prevailing approach, LoRA, models updates using a low-rank decomposition, but its expressivity is inherently constrained by the rank. Recent methods like HiRA aim to increase expressivity by incorporating a Hadamard product with the frozen weights, but still rely on the structure of the pre-trained model. We introduce ABBA, a new PEFT architecture that reparameterizes the update as a Hadamard product of two independently learnable low-rank matrices. In contrast to prior work, ABBA fully decouples the update from the pre-trained weights, enabling both components to be optimized freely. This leads to significantly higher expressivity under the same parameter budget. We formally analyze ABBA's expressive capacity and validate its advantages through matrix reconstruction experiments. Empirically, ABBA achieves state-of-the-art results on arithmetic and commonsense reasoning benchmarks, consistently outperforming existing PEFT methods by a significant margin across multiple models. Our code is publicly available at: https://github.com/CERT-Lab/abba.
Regularization-based Framework for Quantization-, Fault- and Variability-Aware Training
Mar 05, 2025Abstract:Efficient inference is critical for deploying deep learning models on edge AI devices. Low-bit quantization (e.g., 3- and 4-bit) with fixed-point arithmetic improves efficiency, while low-power memory technologies like analog nonvolatile memory enable further gains. However, these methods introduce non-ideal hardware behavior, including bit faults and device-to-device variability. We propose a regularization-based quantization-aware training (QAT) framework that supports fixed, learnable step-size, and learnable non-uniform quantization, achieving competitive results on CIFAR-10 and ImageNet. Our method also extends to Spiking Neural Networks (SNNs), demonstrating strong performance on 4-bit networks on CIFAR10-DVS and N-Caltech 101. Beyond quantization, our framework enables fault and variability-aware fine-tuning, mitigating stuck-at faults (fixed weight bits) and device resistance variability. Compared to prior fault-aware training, our approach significantly improves performance recovery under upto 20% bit-fault rate and 40% device-to-device variability. Our results establish a generalizable framework for quantization and robustness-aware training, enhancing efficiency and reliability in low-power, non-ideal hardware.
Fed-SB: A Silver Bullet for Extreme Communication Efficiency and Performance in (Private) Federated LoRA Fine-Tuning
Feb 21, 2025Abstract:Low-Rank Adaptation (LoRA) has become ubiquitous for efficiently fine-tuning foundation models. However, federated fine-tuning using LoRA is challenging due to suboptimal updates arising from traditional federated averaging of individual adapters. Existing solutions either incur prohibitively high communication cost that scales linearly with the number of clients or suffer from performance degradation due to limited expressivity. We introduce Federated Silver Bullet (Fed-SB), a novel approach for federated fine-tuning of LLMs using LoRA-SB, a recently proposed low-rank adaptation method. LoRA-SB optimally aligns the optimization trajectory with the ideal low-rank full fine-tuning projection by learning a small square matrix (R) between adapters B and A, keeping other components fixed. Direct averaging of R guarantees exact updates, substantially reducing communication cost, which remains independent of the number of clients, and enables scalability. Fed-SB achieves state-of-the-art performance across commonsense reasoning, arithmetic reasoning, and language inference tasks while reducing communication costs by up to 230x. In private settings, Fed-SB further improves performance by (1) reducing trainable parameters, thereby lowering the noise required for differential privacy and (2) avoiding noise amplification introduced by other methods. Overall, Fed-SB establishes a new Pareto frontier in the tradeoff between communication and performance, offering an efficient and scalable solution for both private and non-private federated fine-tuning. Our code is publicly available at https://github.com/CERT-Lab/fed-sb.
Humanity's Last Exam
Jan 24, 2025Abstract:Benchmarks are important tools for tracking the rapid advancements in large language model (LLM) capabilities. However, benchmarks are not keeping pace in difficulty: LLMs now achieve over 90\% accuracy on popular benchmarks like MMLU, limiting informed measurement of state-of-the-art LLM capabilities. In response, we introduce Humanity's Last Exam (HLE), a multi-modal benchmark at the frontier of human knowledge, designed to be the final closed-ended academic benchmark of its kind with broad subject coverage. HLE consists of 3,000 questions across dozens of subjects, including mathematics, humanities, and the natural sciences. HLE is developed globally by subject-matter experts and consists of multiple-choice and short-answer questions suitable for automated grading. Each question has a known solution that is unambiguous and easily verifiable, but cannot be quickly answered via internet retrieval. State-of-the-art LLMs demonstrate low accuracy and calibration on HLE, highlighting a significant gap between current LLM capabilities and the expert human frontier on closed-ended academic questions. To inform research and policymaking upon a clear understanding of model capabilities, we publicly release HLE at https://lastexam.ai.
A General Framework for Inference-time Scaling and Steering of Diffusion Models
Jan 16, 2025Abstract:Diffusion models produce impressive results in modalities ranging from images and video to protein design and text. However, generating samples with user-specified properties remains a challenge. Recent research proposes fine-tuning models to maximize rewards that capture desired properties, but these methods require expensive training and are prone to mode collapse. In this work, we propose Feynman Kac (FK) steering, an inference-time framework for steering diffusion models with reward functions. FK steering works by sampling a system of multiple interacting diffusion processes, called particles, and resampling particles at intermediate steps based on scores computed using functions called potentials. Potentials are defined using rewards for intermediate states and are selected such that a high value indicates that the particle will yield a high-reward sample. We explore various choices of potentials, intermediate rewards, and samplers. We evaluate FK steering on text-to-image and text diffusion models. For steering text-to-image models with a human preference reward, we find that FK steering a 0.8B parameter model outperforms a 2.6B parameter fine-tuned model on prompt fidelity, with faster sampling and no training. For steering text diffusion models with rewards for text quality and specific text attributes, we find that FK steering generates lower perplexity, more linguistically acceptable outputs and enables gradient-free control of attributes like toxicity. Our results demonstrate that inference-time scaling and steering of diffusion models, even with off-the-shelf rewards, can provide significant sample quality gains and controllability benefits. Code is available at https://github.com/zacharyhorvitz/Fk-Diffusion-Steering .
Initialization using Update Approximation is a Silver Bullet for Extremely Efficient Low-Rank Fine-Tuning
Nov 29, 2024



Abstract:Low-rank adapters have become a standard approach for efficiently fine-tuning large language models (LLMs), but they often fall short of achieving the performance of full fine-tuning. We propose a method, LoRA Silver Bullet or LoRA-SB, that approximates full fine-tuning within low-rank subspaces using a carefully designed initialization strategy. We theoretically demonstrate that the architecture of LoRA-XS, which inserts a trainable (r x r) matrix between B and A while keeping other matrices fixed, provides the precise conditions needed for this approximation. We leverage its constrained update space to achieve optimal scaling for high-rank gradient updates while removing the need for hyperparameter tuning. We prove that our initialization offers an optimal low-rank approximation of the initial gradient and preserves update directions throughout training. Extensive experiments across mathematical reasoning, commonsense reasoning, and language understanding tasks demonstrate that our approach exceeds the performance of standard LoRA while using 27-90x fewer parameters, and comprehensively outperforms LoRA-XS. Our findings establish that it is possible to simulate full fine-tuning in low-rank subspaces, and achieve significant efficiency gains without sacrificing performance. Our code is publicly available at https://github.com/RaghavSinghal10/lora-sb.
Temporal and Spatial Reservoir Ensembling Techniques for Liquid State Machines
Nov 18, 2024Abstract:Reservoir computing (RC), is a class of computational methods such as Echo State Networks (ESN) and Liquid State Machines (LSM) describe a generic method to perform pattern recognition and temporal analysis with any non-linear system. This is enabled by Reservoir Computing being a shallow network model with only Input, Reservoir, and Readout layers where input and reservoir weights are not learned (only the readout layer is trained). LSM is a special case of Reservoir computing inspired by the organization of neurons in the brain and generally refers to spike-based Reservoir computing approaches. LSMs have been successfully used to showcase decent performance on some neuromorphic vision and speech datasets but a common problem associated with LSMs is that since the model is more-or-less fixed, the main way to improve the performance is by scaling up the Reservoir size, but that only gives diminishing rewards despite a tremendous increase in model size and computation. In this paper, we propose two approaches for effectively ensembling LSM models - Multi-Length Scale Reservoir Ensemble (MuLRE) and Temporal Excitation Partitioned Reservoir Ensemble (TEPRE) and benchmark them on Neuromorphic-MNIST (N-MNIST), Spiking Heidelberg Digits (SHD), and DVSGesture datasets, which are standard neuromorphic benchmarks. We achieve 98.1% test accuracy on N-MNIST with a 3600-neuron LSM model which is higher than any prior LSM-based approach and 77.8% test accuracy on the SHD dataset which is on par with a standard Recurrent Spiking Neural Network trained by Backprop Through Time (BPTT). We also propose receptive field-based input weights to the Reservoir to work alongside the Multi-Length Scale Reservoir ensemble model for vision tasks. Thus, we introduce effective means of scaling up the performance of LSM models and evaluate them against relevant neuromorphic benchmarks
Exact Aggregation for Federated and Efficient Fine-Tuning of Foundation Models
Oct 12, 2024



Abstract:Low-Rank Adaptation (LoRA) is a popular technique for efficient fine-tuning of foundation models. However, applying LoRA in federated learning environments, where data is distributed across multiple clients, presents unique challenges. Existing methods rely on traditional federated averaging of LoRA adapters, resulting in inexact updates. To address this, we propose Federated Exact LoRA, or FedEx-LoRA, which adds a residual error term to the pretrained frozen weight matrix. Our approach achieves exact updates with minimal computational and communication overhead, preserving LoRA's efficiency. We evaluate the method on various Natural Language Understanding (NLU) and Natural Language Generation (NLG) tasks, showing consistent performance gains over state-of-the-art methods across multiple settings. Through extensive analysis, we quantify that the deviations in updates from the ideal solution are significant, highlighting the need for exact aggregation. Our method's simplicity, efficiency, and broad applicability position it as a promising solution for accurate and effective federated fine-tuning of foundation models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge