Nikhil Cherian Kurian
PathoGen-X: A Cross-Modal Genomic Feature Trans-Align Network for Enhanced Survival Prediction from Histopathology Images
Nov 01, 2024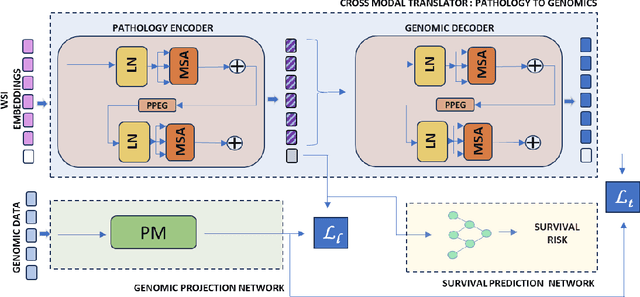
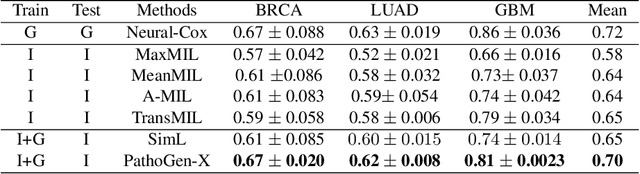
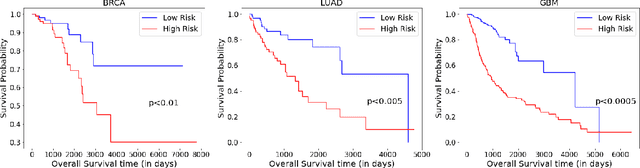
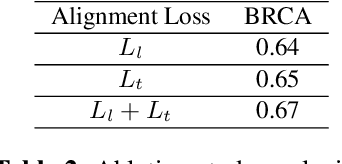
Abstract:Accurate survival prediction is essential for personalized cancer treatment. However, genomic data - often a more powerful predictor than pathology data - is costly and inaccessible. We present the cross-modal genomic feature translation and alignment network for enhanced survival prediction from histopathology images (PathoGen-X). It is a deep learning framework that leverages both genomic and imaging data during training, relying solely on imaging data at testing. PathoGen-X employs transformer-based networks to align and translate image features into the genomic feature space, enhancing weaker imaging signals with stronger genomic signals. Unlike other methods, PathoGen-X translates and aligns features without projecting them to a shared latent space and requires fewer paired samples. Evaluated on TCGA-BRCA, TCGA-LUAD, and TCGA-GBM datasets, PathoGen-X demonstrates strong survival prediction performance, emphasizing the potential of enriched imaging models for accessible cancer prognosis.
Efficient Quality Control of Whole Slide Pathology Images with Human-in-the-loop Training
Sep 29, 2024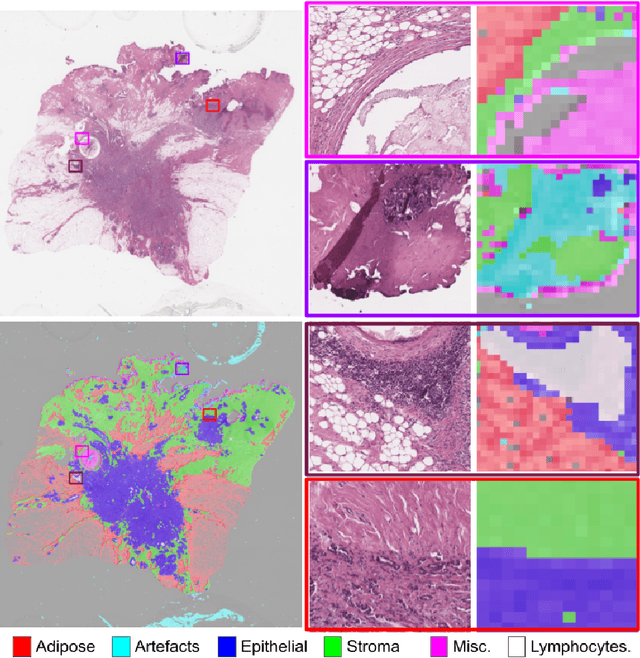
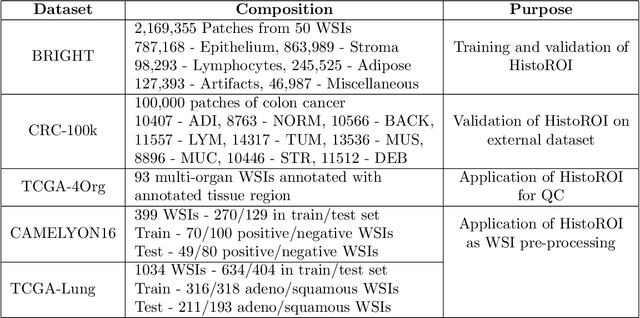
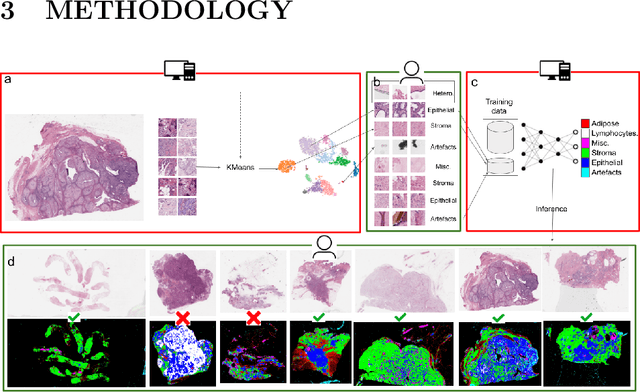
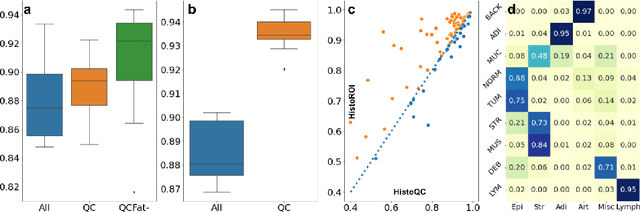
Abstract:Histopathology whole slide images (WSIs) are being widely used to develop deep learning-based diagnostic solutions, especially for precision oncology. Most of these diagnostic softwares are vulnerable to biases and impurities in the training and test data which can lead to inaccurate diagnoses. For instance, WSIs contain multiple types of tissue regions, at least some of which might not be relevant to the diagnosis. We introduce HistoROI, a robust yet lightweight deep learning-based classifier to segregate WSI into six broad tissue regions -- epithelium, stroma, lymphocytes, adipose, artifacts, and miscellaneous. HistoROI is trained using a novel human-in-the-loop and active learning paradigm that ensures variations in training data for labeling-efficient generalization. HistoROI consistently performs well across multiple organs, despite being trained on only a single dataset, demonstrating strong generalization. Further, we have examined the utility of HistoROI in improving the performance of downstream deep learning-based tasks using the CAMELYON breast cancer lymph node and TCGA lung cancer datasets. For the former dataset, the area under the receiver operating characteristic curve (AUC) for metastasis versus normal tissue of a neural network trained using weakly supervised learning increased from 0.88 to 0.92 by filtering the data using HistoROI. Similarly, the AUC increased from 0.88 to 0.93 for the classification between adenocarcinoma and squamous cell carcinoma on the lung cancer dataset. We also found that the performance of the HistoROI improves upon HistoQC for artifact detection on a test dataset of 93 annotated WSIs. The limitations of the proposed model are analyzed, and potential extensions are also discussed.
* 18 pages
Heterogeneous graphs model spatial relationships between biological entities for breast cancer diagnosis
Jul 16, 2023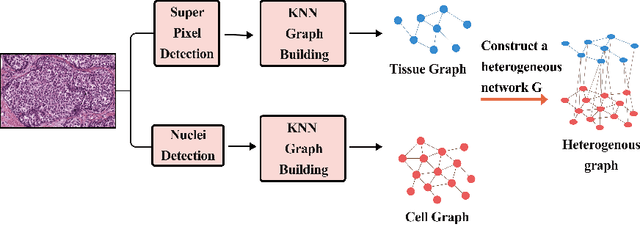
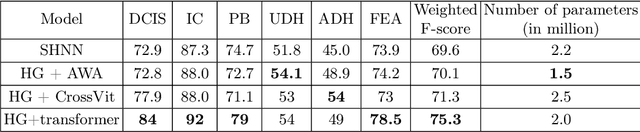
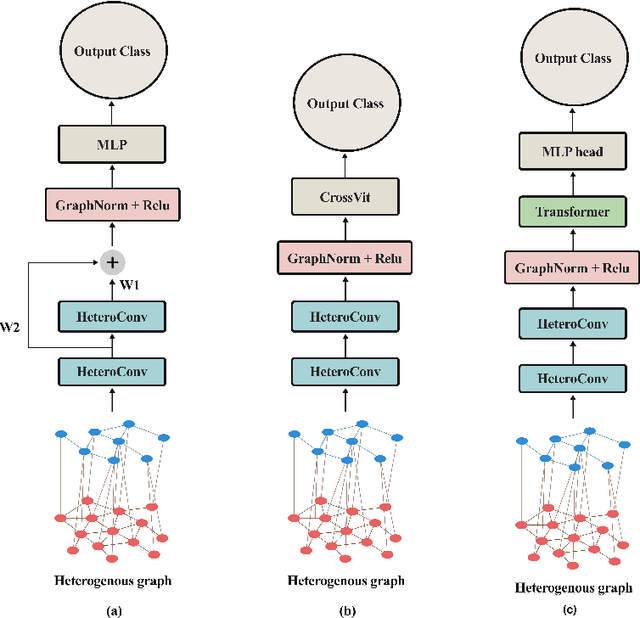
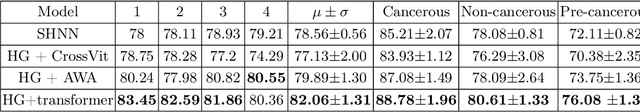
Abstract:The heterogeneity of breast cancer presents considerable challenges for its early detection, prognosis, and treatment selection. Convolutional neural networks often neglect the spatial relationships within histopathological images, which can limit their accuracy. Graph neural networks (GNNs) offer a promising solution by coding the spatial relationships within images. Prior studies have investigated the modeling of histopathological images as cell and tissue graphs, but they have not fully tapped into the potential of extracting interrelationships between these biological entities. In this paper, we present a novel approach using a heterogeneous GNN that captures the spatial and hierarchical relations between cell and tissue graphs to enhance the extraction of useful information from histopathological images. We also compare the performance of a cross-attention-based network and a transformer architecture for modeling the intricate relationships within tissue and cell graphs. Our model demonstrates superior efficiency in terms of parameter count and achieves higher accuracy compared to the transformer-based state-of-the-art approach on three publicly available breast cancer datasets -- BRIGHT, BreakHis, and BACH.
Robust Semi-Supervised Learning for Histopathology Images through Self-Supervision Guided Out-of-Distribution Scoring
Mar 17, 2023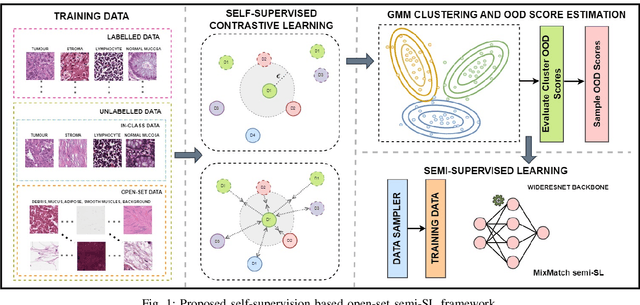
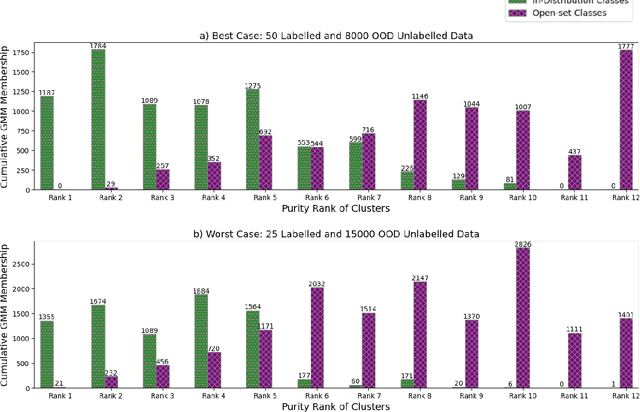
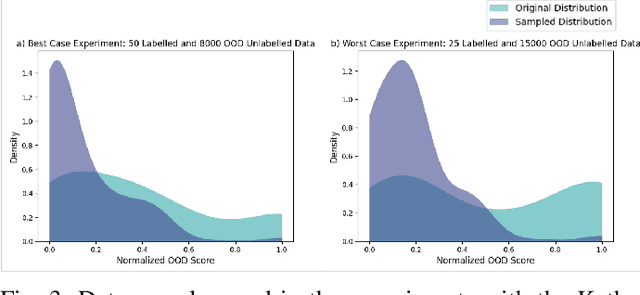
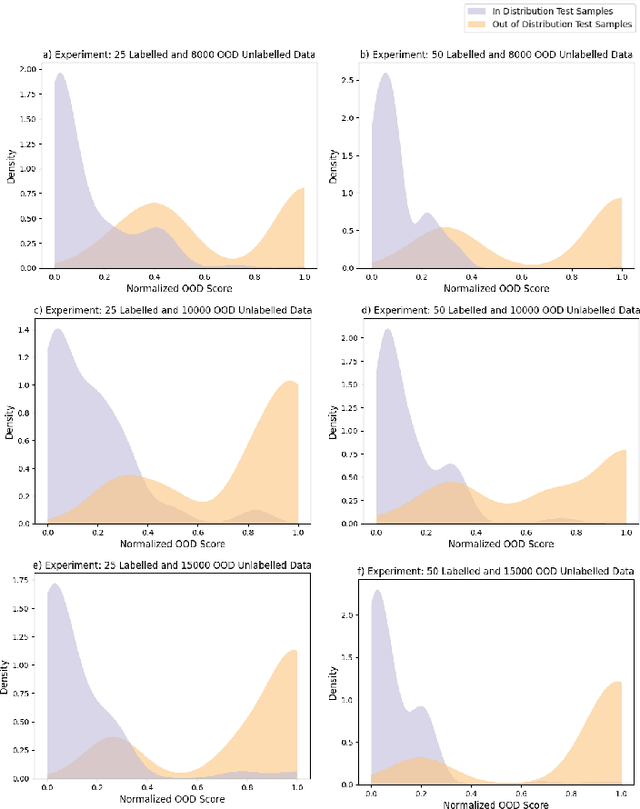
Abstract:Semi-supervised learning (semi-SL) is a promising alternative to supervised learning for medical image analysis when obtaining good quality supervision for medical imaging is difficult. However, semi-SL assumes that the underlying distribution of unaudited data matches that of the few labeled samples, which is often violated in practical settings, particularly in medical images. The presence of out-of-distribution (OOD) samples in the unlabeled training pool of semi-SL is inevitable and can reduce the efficiency of the algorithm. Common preprocessing methods to filter out outlier samples may not be suitable for medical images that involve a wide range of anatomical structures and rare morphologies. In this paper, we propose a novel pipeline for addressing open-set supervised learning challenges in digital histology images. Our pipeline efficiently estimates an OOD score for each unlabelled data point based on self-supervised learning to calibrate the knowledge needed for a subsequent semi-SL framework. The outlier score derived from the OOD detector is used to modulate sample selection for the subsequent semi-SL stage, ensuring that samples conforming to the distribution of the few labeled samples are more frequently exposed to the subsequent semi-SL framework. Our framework is compatible with any semi-SL framework, and we base our experiments on the popular Mixmatch semi-SL framework. We conduct extensive studies on two digital pathology datasets, Kather colorectal histology dataset and a dataset derived from TCGA-BRCA whole slide images, and establish the effectiveness of our method by comparing with popular methods and frameworks in semi-SL algorithms through various experiments.
Magnification Invariant Medical Image Analysis: A Comparison of Convolutional Networks, Vision Transformers, and Token Mixers
Feb 22, 2023Abstract:Convolution Neural Networks (CNNs) are widely used in medical image analysis, but their performance degrade when the magnification of testing images differ from the training images. The inability of CNNs to generalize across magnification scales can result in sub-optimal performance on external datasets. This study aims to evaluate the robustness of various deep learning architectures in the analysis of breast cancer histopathological images with varying magnification scales at training and testing stages. Here we explore and compare the performance of multiple deep learning architectures, including CNN-based ResNet and MobileNet, self-attention-based Vision Transformers and Swin Transformers, and token-mixing models, such as FNet, ConvMixer, MLP-Mixer, and WaveMix. The experiments are conducted using the BreakHis dataset, which contains breast cancer histopathological images at varying magnification levels. We show that performance of WaveMix is invariant to the magnification of training and testing data and can provide stable and good classification accuracy. These evaluations are critical in identifying deep learning architectures that can robustly handle changes in magnification scale, ensuring that scale changes across anatomical structures do not disturb the inference results.
Improving Mitosis Detection Via UNet-based Adversarial Domain Homogenizer
Sep 15, 2022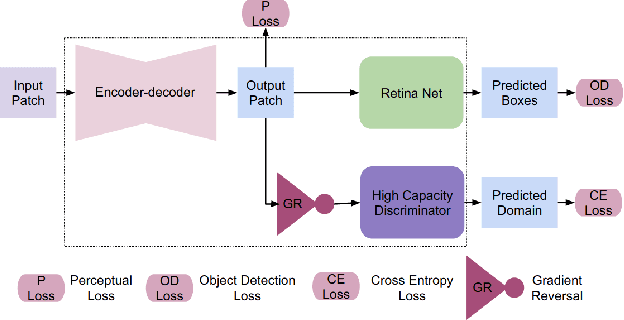

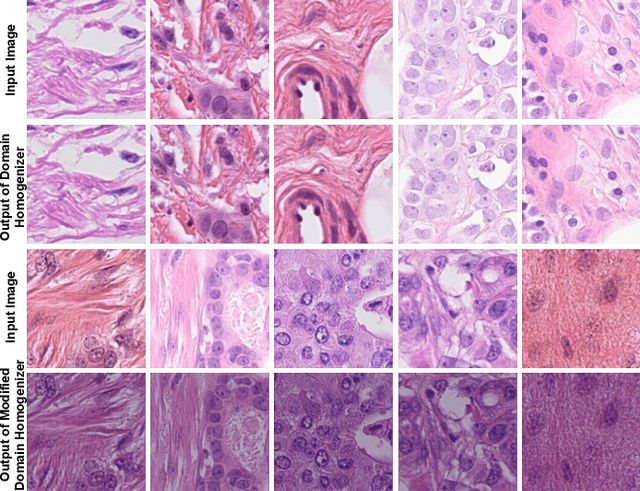
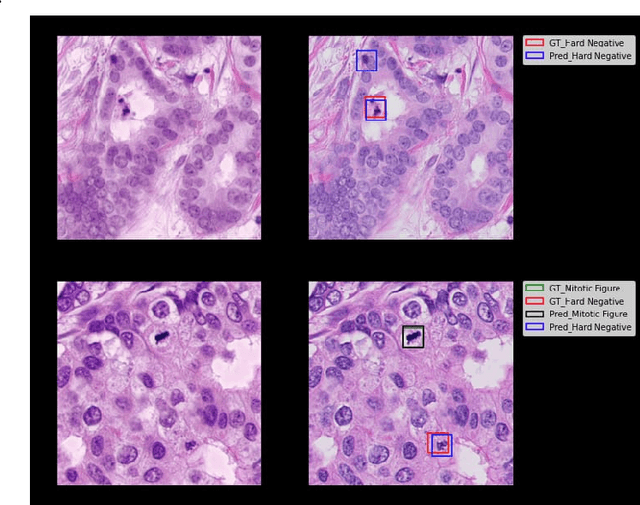
Abstract:The effective localization of mitosis is a critical precursory task for deciding tumor prognosis and grade. Automated mitosis detection through deep learning-oriented image analysis often fails on unseen patient data due to inherent domain biases. This paper proposes a domain homogenizer for mitosis detection that attempts to alleviate domain differences in histology images via adversarial reconstruction of input images. The proposed homogenizer is based on a U-Net architecture and can effectively reduce domain differences commonly seen with histology imaging data. We demonstrate our domain homogenizer's effectiveness by observing the reduction in domain differences between the preprocessed images. Using this homogenizer, along with a subsequent retina-net object detector, we were able to outperform the baselines of the 2021 MIDOG challenge in terms of average precision of the detected mitotic figures.
EGFR Mutation Prediction of Lung Biopsy Images using Deep Learning
Aug 26, 2022
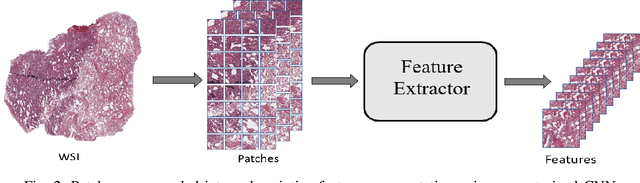

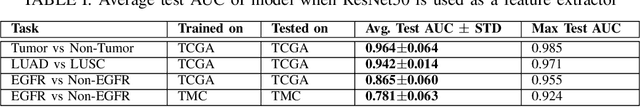
Abstract:The standard diagnostic procedures for targeted therapies in lung cancer treatment involve histological subtyping and subsequent detection of key driver mutations, such as EGFR. Even though molecular profiling can uncover the driver mutation, the process is often expensive and time-consuming. Deep learning-oriented image analysis offers a more economical alternative for discovering driver mutations directly from whole slide images (WSIs). In this work, we used customized deep learning pipelines with weak supervision to identify the morphological correlates of EGFR mutation from hematoxylin and eosin-stained WSIs, in addition to detecting tumor and histologically subtyping it. We demonstrate the effectiveness of our pipeline by conducting rigorous experiments and ablation studies on two lung cancer datasets - TCGA and a private dataset from India. With our pipeline, we achieved an average area under the curve (AUC) of 0.964 for tumor detection, and 0.942 for histological subtyping between adenocarcinoma and squamous cell carcinoma on the TCGA dataset. For EGFR detection, we achieved an average AUC of 0.864 on the TCGA dataset and 0.783 on the dataset from India. Our key learning points include the following. Firstly, there is no particular advantage of using a feature extractor layers trained on histology, if one is going to fine-tune the feature extractor on the target dataset. Secondly, selecting patches with high cellularity, presumably capturing tumor regions, is not always helpful, as the sign of a disease class may be present in the tumor-adjacent stroma.
Deep Multi-Scale U-Net Architecture and Noise-Robust Training Strategies for Histopathological Image Segmentation
May 03, 2022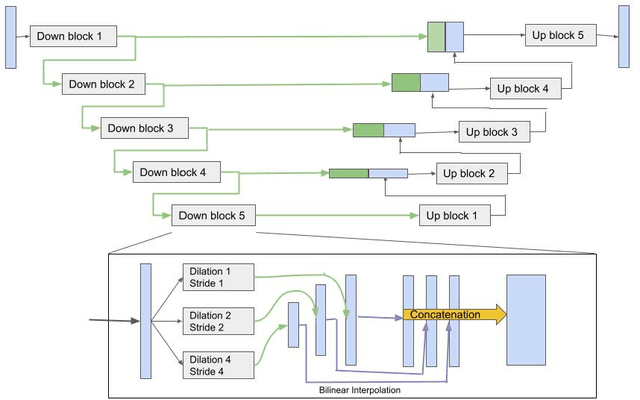
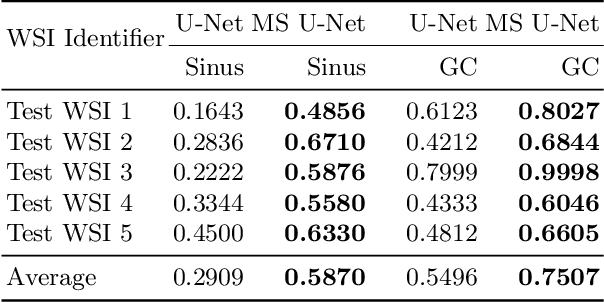

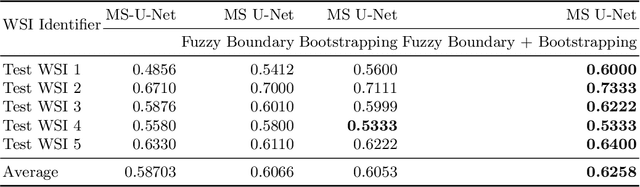
Abstract:Although the U-Net architecture has been extensively used for segmentation of medical images, we address two of its shortcomings in this work. Firstly, the accuracy of vanilla U-Net degrades when the target regions for segmentation exhibit significant variations in shape and size. Even though the U-Net already possesses some capability to analyze features at various scales, we propose to explicitly add multi-scale feature maps in each convolutional module of the U-Net encoder to improve segmentation of histology images. Secondly, the accuracy of a U-Net model also suffers when the annotations for supervised learning are noisy or incomplete. This can happen due to the inherent difficulty for a human expert to identify and delineate all instances of specific pathology very precisely and accurately. We address this challenge by introducing auxiliary confidence maps that emphasize less on the boundaries of the given target regions. Further, we utilize the bootstrapping properties of the deep network to address the missing annotation problem intelligently. In our experiments on a private dataset of breast cancer lymph nodes, where the primary task was to segment germinal centres and sinus histiocytosis, we observed substantial improvement over a U-Net baseline based on the two proposed augmentations.
WSSAMNet: Weakly Supervised Semantic Attentive Medical Image Registration Network
Mar 05, 2022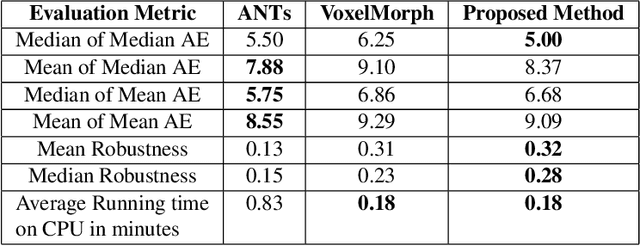
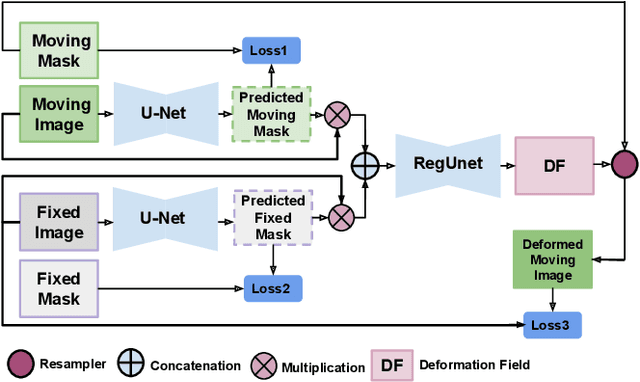
Abstract:We present WSSAMNet, a weakly supervised method for medical image registration. Ours is a two step method, with the first step being the computation of segmentation masks of the fixed and moving volumes. These masks are then used to attend to the input volume, which are then provided as inputs to a registration network in the second step. The registration network computes the deformation field to perform the alignment between the fixed and the moving volumes. We study the effectiveness of our technique on the BraTSReg challenge data against ANTs and VoxelMorph, where we demonstrate that our method performs competitively.
Fast, Self Supervised, Fully Convolutional Color Normalization of H&E Stained Images
Nov 30, 2020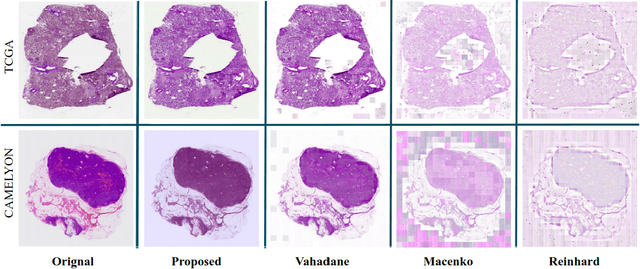
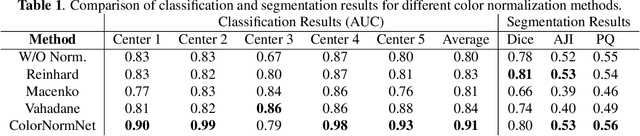

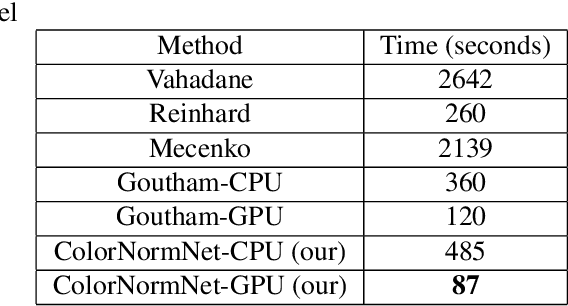
Abstract:Performance of deep learning algorithms decreases drastically if the data distributions of the training and testing sets are different. Due to variations in staining protocols, reagent brands, and habits of technicians, color variation in digital histopathology images is quite common. Color variation causes problems for the deployment of deep learning-based solutions for automatic diagnosis system in histopathology. Previously proposed color normalization methods consider a small patch as a reference for normalization, which creates artifacts on out-of-distribution source images. These methods are also slow as most of the computation is performed on CPUs instead of the GPUs. We propose a color normalization technique, which is fast during its self-supervised training as well as inference. Our method is based on a lightweight fully-convolutional neural network and can be easily attached to a deep learning-based pipeline as a pre-processing block. For classification and segmentation tasks on CAMELYON17 and MoNuSeg datasets respectively, the proposed method is faster and gives a greater increase in accuracy than the state of the art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge