Swati Meena
Deep Multi-Scale U-Net Architecture and Noise-Robust Training Strategies for Histopathological Image Segmentation
May 03, 2022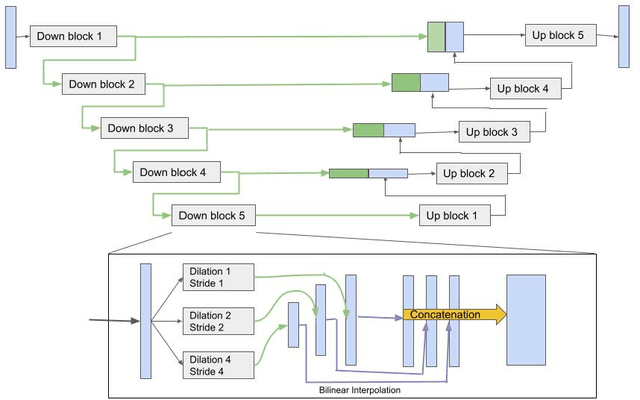
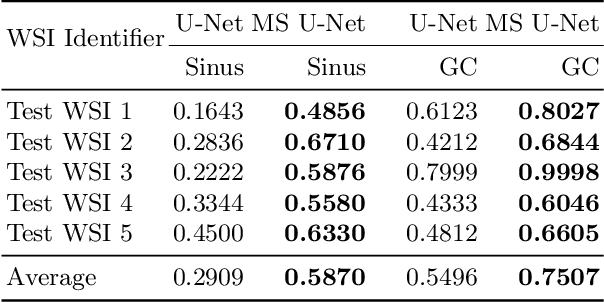

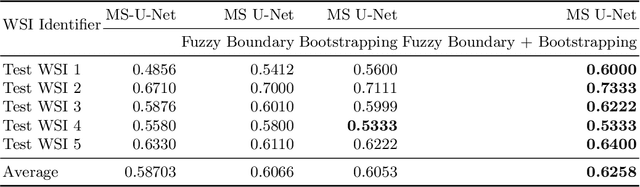
Abstract:Although the U-Net architecture has been extensively used for segmentation of medical images, we address two of its shortcomings in this work. Firstly, the accuracy of vanilla U-Net degrades when the target regions for segmentation exhibit significant variations in shape and size. Even though the U-Net already possesses some capability to analyze features at various scales, we propose to explicitly add multi-scale feature maps in each convolutional module of the U-Net encoder to improve segmentation of histology images. Secondly, the accuracy of a U-Net model also suffers when the annotations for supervised learning are noisy or incomplete. This can happen due to the inherent difficulty for a human expert to identify and delineate all instances of specific pathology very precisely and accurately. We address this challenge by introducing auxiliary confidence maps that emphasize less on the boundaries of the given target regions. Further, we utilize the bootstrapping properties of the deep network to address the missing annotation problem intelligently. In our experiments on a private dataset of breast cancer lymph nodes, where the primary task was to segment germinal centres and sinus histiocytosis, we observed substantial improvement over a U-Net baseline based on the two proposed augmentations.
Breast Cancer Histopathology Image Classification and Localization using Multiple Instance Learning
Feb 16, 2020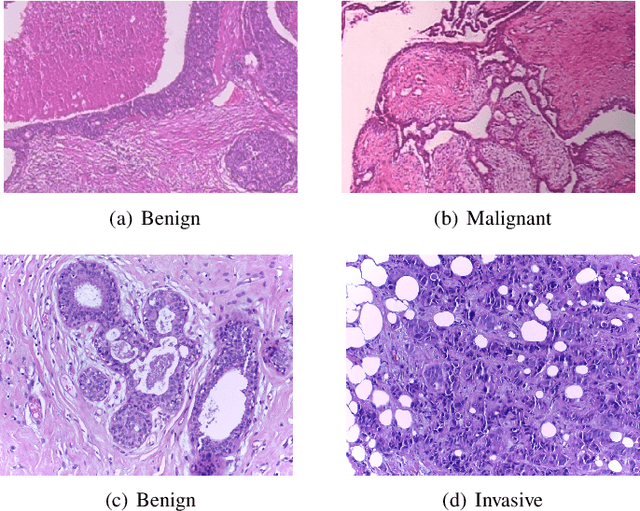
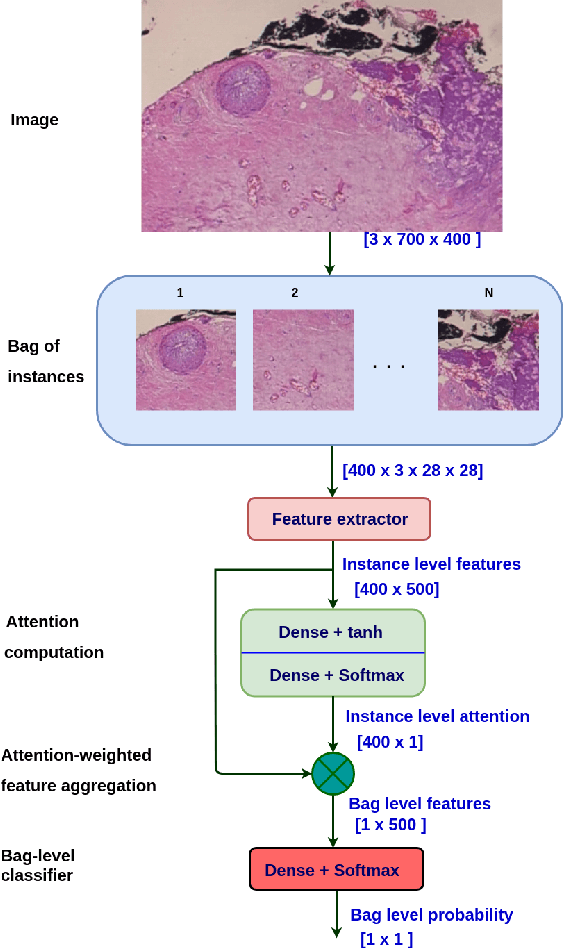
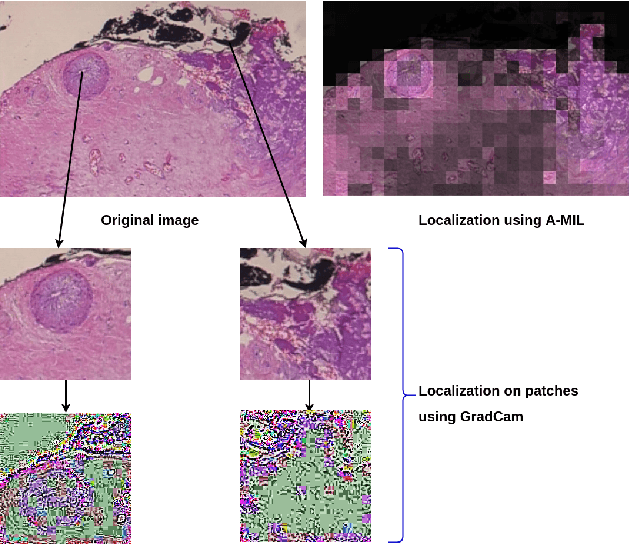
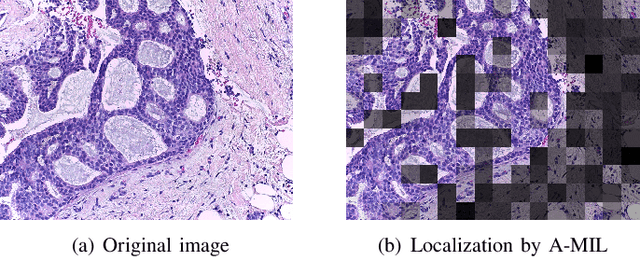
Abstract:Breast cancer has the highest mortality among cancers in women. Computer-aided pathology to analyze microscopic histopathology images for diagnosis with an increasing number of breast cancer patients can bring the cost and delays of diagnosis down. Deep learning in histopathology has attracted attention over the last decade of achieving state-of-the-art performance in classification and localization tasks. The convolutional neural network, a deep learning framework, provides remarkable results in tissue images analysis, but lacks in providing interpretation and reasoning behind the decisions. We aim to provide a better interpretation of classification results by providing localization on microscopic histopathology images. We frame the image classification problem as weakly supervised multiple instance learning problem where an image is collection of patches i.e. instances. Attention-based multiple instance learning (A-MIL) learns attention on the patches from the image to localize the malignant and normal regions in an image and use them to classify the image. We present classification and localization results on two publicly available BreakHIS and BACH dataset. The classification and visualization results are compared with other recent techniques. The proposed method achieves better localization results without compromising classification accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge