Fast, Self Supervised, Fully Convolutional Color Normalization of H&E Stained Images
Paper and Code
Nov 30, 2020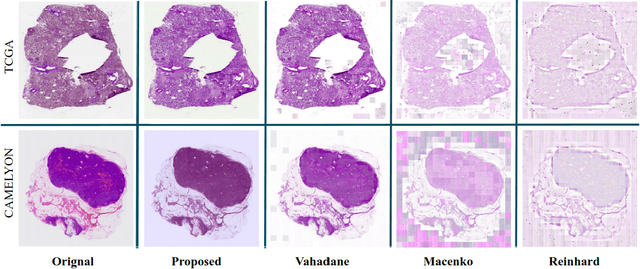
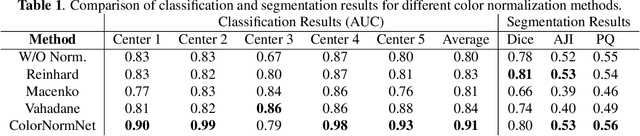

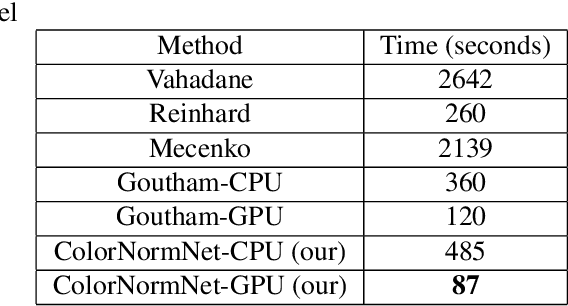
Performance of deep learning algorithms decreases drastically if the data distributions of the training and testing sets are different. Due to variations in staining protocols, reagent brands, and habits of technicians, color variation in digital histopathology images is quite common. Color variation causes problems for the deployment of deep learning-based solutions for automatic diagnosis system in histopathology. Previously proposed color normalization methods consider a small patch as a reference for normalization, which creates artifacts on out-of-distribution source images. These methods are also slow as most of the computation is performed on CPUs instead of the GPUs. We propose a color normalization technique, which is fast during its self-supervised training as well as inference. Our method is based on a lightweight fully-convolutional neural network and can be easily attached to a deep learning-based pipeline as a pre-processing block. For classification and segmentation tasks on CAMELYON17 and MoNuSeg datasets respectively, the proposed method is faster and gives a greater increase in accuracy than the state of the art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge