Amruta Parulekar
AMPS: ASR with Multimodal Paraphrase Supervision
Nov 27, 2024Abstract:Spontaneous or conversational multilingual speech presents many challenges for state-of-the-art automatic speech recognition (ASR) systems. In this work, we present a new technique AMPS that augments a multilingual multimodal ASR system with paraphrase-based supervision for improved conversational ASR in multiple languages, including Hindi, Marathi, Malayalam, Kannada, and Nyanja. We use paraphrases of the reference transcriptions as additional supervision while training the multimodal ASR model and selectively invoke this paraphrase objective for utterances with poor ASR performance. Using AMPS with a state-of-the-art multimodal model SeamlessM4T, we obtain significant relative reductions in word error rates (WERs) of up to 5%. We present detailed analyses of our system using both objective and human evaluation metrics.
PathoGen-X: A Cross-Modal Genomic Feature Trans-Align Network for Enhanced Survival Prediction from Histopathology Images
Nov 01, 2024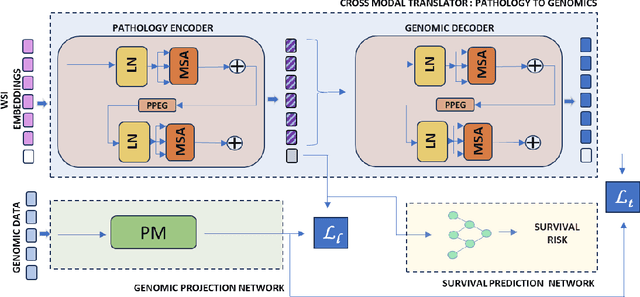
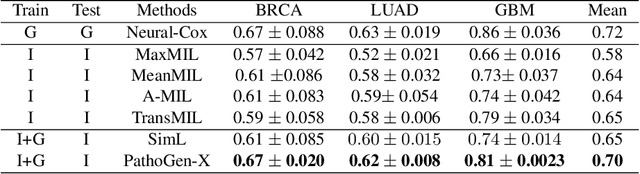
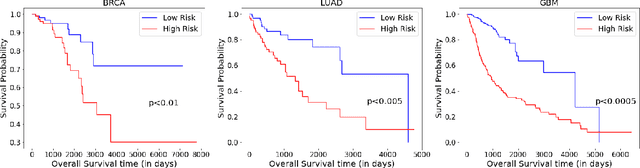
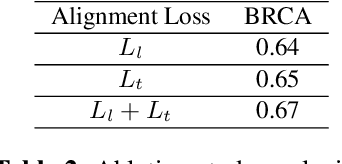
Abstract:Accurate survival prediction is essential for personalized cancer treatment. However, genomic data - often a more powerful predictor than pathology data - is costly and inaccessible. We present the cross-modal genomic feature translation and alignment network for enhanced survival prediction from histopathology images (PathoGen-X). It is a deep learning framework that leverages both genomic and imaging data during training, relying solely on imaging data at testing. PathoGen-X employs transformer-based networks to align and translate image features into the genomic feature space, enhancing weaker imaging signals with stronger genomic signals. Unlike other methods, PathoGen-X translates and aligns features without projecting them to a shared latent space and requires fewer paired samples. Evaluated on TCGA-BRCA, TCGA-LUAD, and TCGA-GBM datasets, PathoGen-X demonstrates strong survival prediction performance, emphasizing the potential of enriched imaging models for accessible cancer prognosis.
Parameter-efficient Adaptation of Multilingual Multimodal Models for Low-resource ASR
Oct 17, 2024



Abstract:Automatic speech recognition (ASR) for low-resource languages remains a challenge due to the scarcity of labeled training data. Parameter-efficient fine-tuning and text-only adaptation are two popular methods that have been used to address such low-resource settings. In this work, we investigate how these techniques can be effectively combined using a multilingual multimodal model like SeamlessM4T. Multimodal models are able to leverage unlabeled text via text-only adaptation with further parameter-efficient ASR fine-tuning, thus boosting ASR performance. We also show cross-lingual transfer from a high-resource language, achieving up to a relative 17% WER reduction over a baseline in a zero-shot setting without any labeled speech.
Combining Datasets with Different Label Sets for Improved Nucleus Segmentation and Classification
Oct 05, 2023



Abstract:Segmentation and classification of cell nuclei in histopathology images using deep neural networks (DNNs) can save pathologists' time for diagnosing various diseases, including cancers, by automating cell counting and morphometric assessments. It is now well-known that the accuracy of DNNs increases with the sizes of annotated datasets available for training. Although multiple datasets of histopathology images with nuclear annotations and class labels have been made publicly available, the set of class labels differ across these datasets. We propose a method to train DNNs for instance segmentation and classification on multiple datasets where the set of classes across the datasets are related but not the same. Specifically, our method is designed to utilize a coarse-to-fine class hierarchy, where the set of classes labeled and annotated in a dataset can be at any level of the hierarchy, as long as the classes are mutually exclusive. Within a dataset, the set of classes need not even be at the same level of the class hierarchy tree. Our results demonstrate that segmentation and classification metrics for the class set used by the test split of a dataset can improve by pre-training on another dataset that may even have a different set of classes due to the expansion of the training set enabled by our method. Furthermore, generalization to previously unseen datasets also improves by combining multiple other datasets with different sets of classes for training. The improvement is both qualitative and quantitative. The proposed method can be adapted for various loss functions, DNN architectures, and application domains.
Artificial Intelligence-based Eosinophil Counting in Gastrointestinal Biopsies
Nov 25, 2022Abstract:Normally eosinophils are present in the gastrointestinal (GI) tract of healthy individuals. When the eosinophils increase beyond their usual amount in the GI tract, a patient gets varied symptoms. Clinicians find it difficult to diagnose this condition called eosinophilia. Early diagnosis can help in treating patients. Histopathology is the gold standard in the diagnosis for this condition. As this is an under-diagnosed condition, counting eosinophils in the GI tract biopsies is important. In this study, we trained and tested a deep neural network based on UNet to detect and count eosinophils in GI tract biopsies. We used connected component analysis to extract the eosinophils. We studied correlation of eosinophilic infiltration counted by AI with a manual count. GI tract biopsy slides were stained with H&E stain. Slides were scanned using a camera attached to a microscope and five high-power field images were taken per slide. Pearson correlation coefficient was 85% between the machine-detected and manual eosinophil counts on 300 held-out (test) images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge