Pranav Jeevan
Indian Institute of Technology Bombay
Spatially-Aware Mixture of Experts with Log-Logistic Survival Modeling for Whole-Slide Images
Nov 17, 2025Abstract:Accurate survival prediction from histopathology whole-slide images (WSIs) remains challenging due to their gigapixel resolution, strong spatial heterogeneity, and complex survival distributions. We introduce a comprehensive computational pathology framework that addresses these limitations through four complementary innovations: (1) Quantile-Gated Patch Selection for dynamically identifying prognostically relevant regions, (2) Graph-Guided Clustering to group patches by spatial and morphological similarity, (3) Hierarchical Context Attention to model both local tissue interactions and global slide-level context, and (4) an Expert-Driven Mixture of Log-Logistics module that flexibly models complex survival distributions. Across large TCGA cohorts, our method achieves state-of-the-art performance, yielding time-dependent concordance indices of 0.644 on LUAD, 0.751 on KIRC, and 0.752 on BRCA, consistently outperforming both histology-only and multimodal baselines. The framework further provides improved calibration and interpretability, advancing the use of WSIs for personalized cancer prognosis.
Survival Modeling from Whole Slide Images via Patch-Level Graph Clustering and Mixture Density Experts
Jul 22, 2025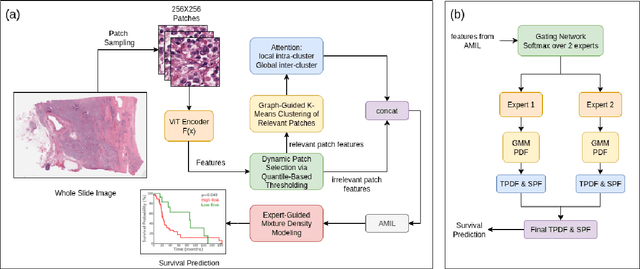
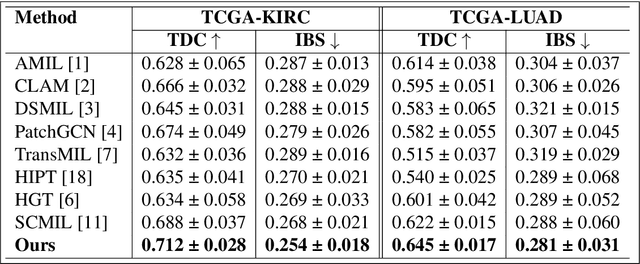
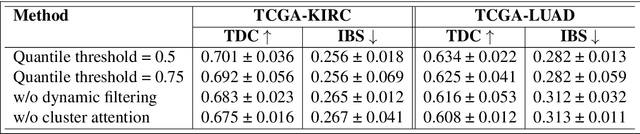
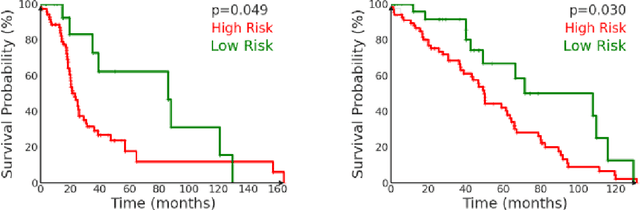
Abstract:We introduce a modular framework for predicting cancer-specific survival from whole slide pathology images (WSIs) that significantly improves upon the state-of-the-art accuracy. Our method integrating four key components. Firstly, to tackle large size of WSIs, we use dynamic patch selection via quantile-based thresholding for isolating prognostically informative tissue regions. Secondly, we use graph-guided k-means clustering to capture phenotype-level heterogeneity through spatial and morphological coherence. Thirdly, we use attention mechanisms that model both intra- and inter-cluster relationships to contextualize local features within global spatial relations between various types of tissue compartments. Finally, we use an expert-guided mixture density modeling for estimating complex survival distributions using Gaussian mixture models. The proposed model achieves a concordance index of $0.712 \pm 0.028$ and Brier score of $0.254 \pm 0.018$ on TCGA-KIRC (renal cancer), and a concordance index of $0.645 \pm 0.017$ and Brier score of $0.281 \pm 0.031$ on TCGA-LUAD (lung adenocarcinoma). These results are significantly better than the state-of-art and demonstrate predictive potential of the proposed method across diverse cancer types.
FLD+: Data-efficient Evaluation Metric for Generative Models
Nov 23, 2024Abstract:We introduce a new metric to assess the quality of generated images that is more reliable, data-efficient, compute-efficient, and adaptable to new domains than the previous metrics, such as Fr\'echet Inception Distance (FID). The proposed metric is based on normalizing flows, which allows for the computation of density (exact log-likelihood) of images from any domain. Thus, unlike FID, the proposed Flow-based Likelihood Distance Plus (FLD+) metric exhibits strongly monotonic behavior with respect to different types of image degradations, including noise, occlusion, diffusion steps, and generative model size. Additionally, because normalizing flow can be trained stably and efficiently, FLD+ achieves stable results with two orders of magnitude fewer images than FID (which requires more images to reliably compute Fr\'echet distance between features of large samples of real and generated images). We made FLD+ computationally even more efficient by applying normalizing flows to features extracted in a lower-dimensional latent space instead of using a pre-trained network. We also show that FLD+ can easily be retrained on new domains, such as medical images, unlike the networks behind previous metrics -- such as InceptionNetV3 pre-trained on ImageNet.
Evaluation Metric for Quality Control and Generative Models in Histopathology Images
Nov 01, 2024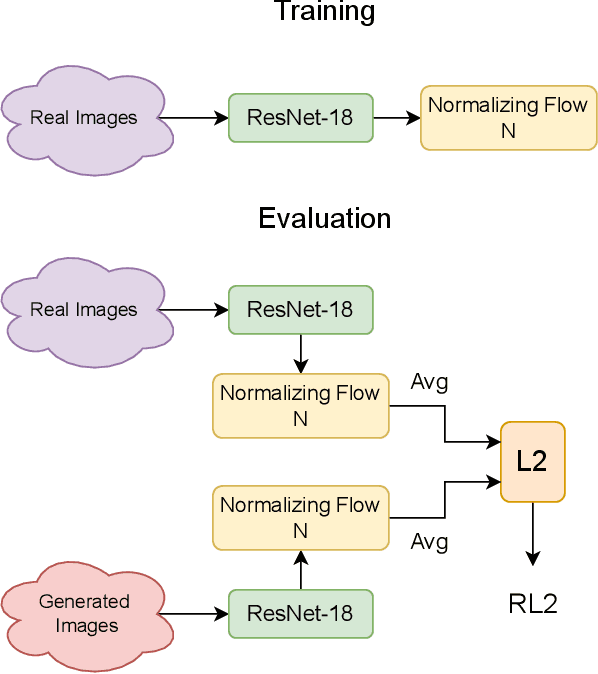
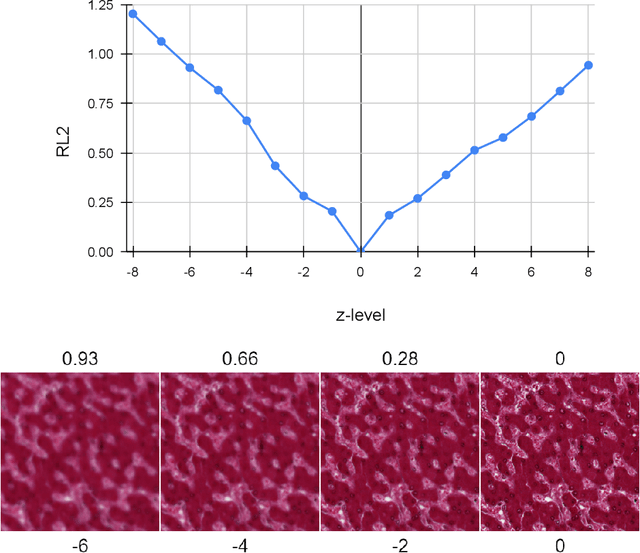
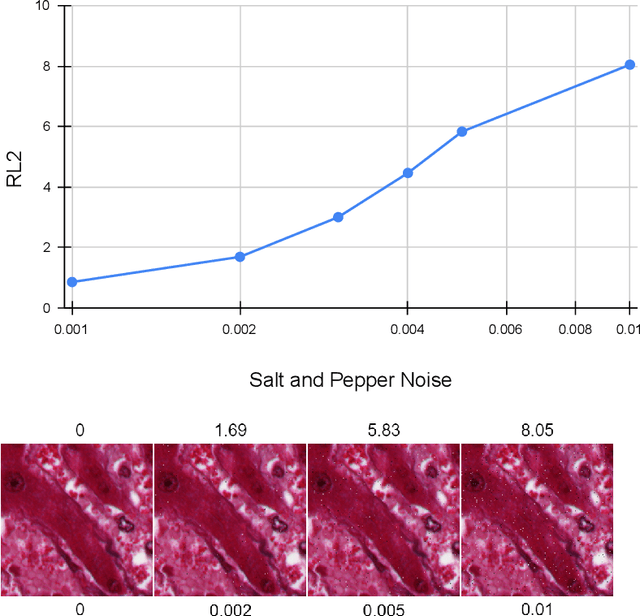
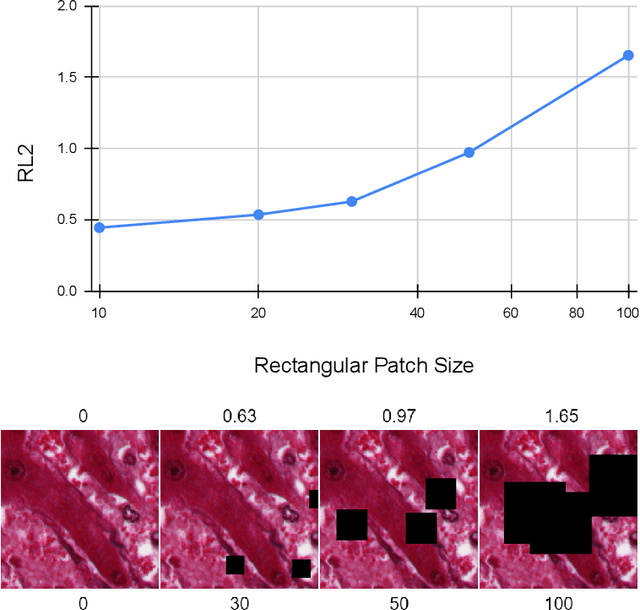
Abstract:Our study introduces ResNet-L2 (RL2), a novel metric for evaluating generative models and image quality in histopathology, addressing limitations of traditional metrics, such as Frechet inception distance (FID), when the data is scarce. RL2 leverages ResNet features with a normalizing flow to calculate RMSE distance in the latent space, providing reliable assessments across diverse histopathology datasets. We evaluated the performance of RL2 on degradation types, such as blur, Gaussian noise, salt-and-pepper noise, and rectangular patches, as well as diffusion processes. RL2's monotonic response to increasing degradation makes it well-suited for models that assess image quality, proving a valuable advancement for evaluating image generation techniques in histopathology. It can also be used to discard low-quality patches while sampling from a whole slide image. It is also significantly lighter and faster compared to traditional metrics and requires fewer images to give stable metric value.
Normalizing Flow Based Metric for Image Generation
Oct 02, 2024



Abstract:We propose two new evaluation metrics to assess realness of generated images based on normalizing flows: a simpler and efficient flow-based likelihood distance (FLD) and a more exact dual-flow based likelihood distance (D-FLD). Because normalizing flows can be used to compute the exact likelihood, the proposed metrics assess how closely generated images align with the distribution of real images from a given domain. This property gives the proposed metrics a few advantages over the widely used Fr\'echet inception distance (FID) and other recent metrics. Firstly, the proposed metrics need only a few hundred images to stabilize (converge in mean), as opposed to tens of thousands needed for FID, and at least a few thousand for the other metrics. This allows confident evaluation of even small sets of generated images, such as validation batches inside training loops. Secondly, the network used to compute the proposed metric has over an order of magnitude fewer parameters compared to Inception-V3 used to compute FID, making it computationally more efficient. For assessing the realness of generated images in new domains (e.g., x-ray images), ideally these networks should be retrained on real images to model their distinct distributions. Thus, our smaller network will be even more advantageous for new domains. Extensive experiments show that the proposed metrics have the desired monotonic relationships with the extent of image degradation of various kinds.
WaveMixSR-V2: Enhancing Super-resolution with Higher Efficiency
Sep 16, 2024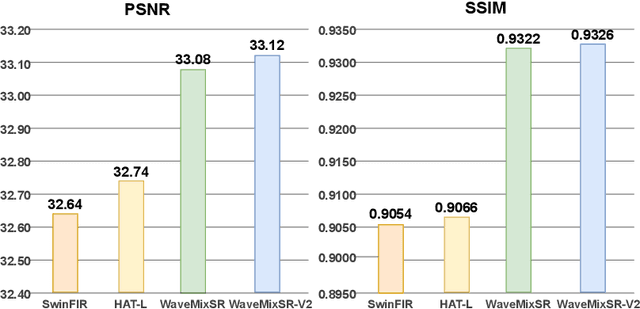
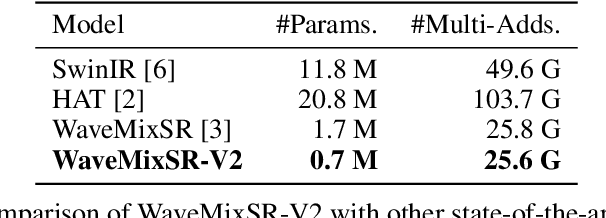
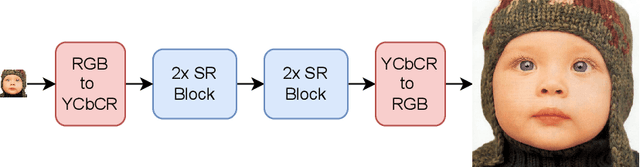
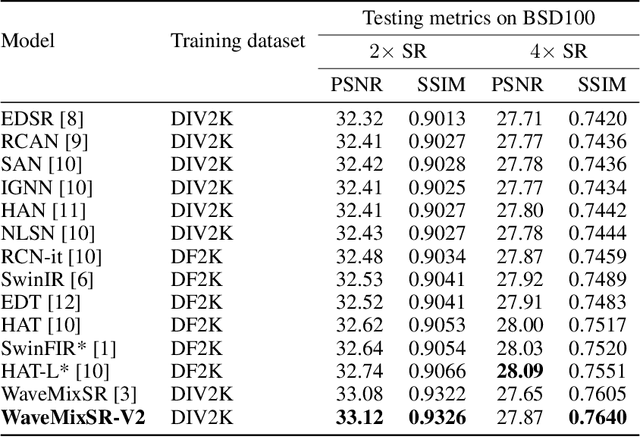
Abstract:Recent advancements in single image super-resolution have been predominantly driven by token mixers and transformer architectures. WaveMixSR utilized the WaveMix architecture, employing a two-dimensional discrete wavelet transform for spatial token mixing, achieving superior performance in super-resolution tasks with remarkable resource efficiency. In this work, we present an enhanced version of the WaveMixSR architecture by (1) replacing the traditional transpose convolution layer with a pixel shuffle operation and (2) implementing a multistage design for higher resolution tasks ($4\times$). Our experiments demonstrate that our enhanced model -- WaveMixSR-V2 -- outperforms other architectures in multiple super-resolution tasks, achieving state-of-the-art for the BSD100 dataset, while also consuming fewer resources, exhibits higher parameter efficiency, lower latency and higher throughput. Our code is available at https://github.com/pranavphoenix/WaveMixSR.
Which Backbone to Use: A Resource-efficient Domain Specific Comparison for Computer Vision
Jun 09, 2024



Abstract:In contemporary computer vision applications, particularly image classification, architectural backbones pre-trained on large datasets like ImageNet are commonly employed as feature extractors. Despite the widespread use of these pre-trained convolutional neural networks (CNNs), there remains a gap in understanding the performance of various resource-efficient backbones across diverse domains and dataset sizes. Our study systematically evaluates multiple lightweight, pre-trained CNN backbones under consistent training settings across a variety of datasets, including natural images, medical images, galaxy images, and remote sensing images. This comprehensive analysis aims to aid machine learning practitioners in selecting the most suitable backbone for their specific problem, especially in scenarios involving small datasets where fine-tuning a pre-trained network is crucial. Even though attention-based architectures are gaining popularity, we observed that they tend to perform poorly under low data finetuning tasks compared to CNNs. We also observed that some CNN architectures such as ConvNeXt, RegNet and EfficientNet performs well compared to others on a diverse set of domains consistently. Our findings provide actionable insights into the performance trade-offs and effectiveness of different backbones, facilitating informed decision-making in model selection for a broad spectrum of computer vision domains. Our code is available here: https://github.com/pranavphoenix/Backbones
Advancing Gene Selection in Oncology: A Fusion of Deep Learning and Sparsity for Precision Gene Selection
Mar 04, 2024



Abstract:Gene selection plays a pivotal role in oncology research for improving outcome prediction accuracy and facilitating cost-effective genomic profiling for cancer patients. This paper introduces two gene selection strategies for deep learning-based survival prediction models. The first strategy uses a sparsity-inducing method while the second one uses importance based gene selection for identifying relevant genes. Our overall approach leverages the power of deep learning to model complex biological data structures, while sparsity-inducing methods ensure the selection process focuses on the most informative genes, minimizing noise and redundancy. Through comprehensive experimentation on diverse genomic and survival datasets, we demonstrate that our strategy not only identifies gene signatures with high predictive power for survival outcomes but can also streamlines the process for low-cost genomic profiling. The implications of this research are profound as it offers a scalable and effective tool for advancing personalized medicine and targeted cancer therapies. By pushing the boundaries of gene selection methodologies, our work contributes significantly to the ongoing efforts in cancer genomics, promising improved diagnostic and prognostic capabilities in clinical settings.
Heterogeneous graphs model spatial relationships between biological entities for breast cancer diagnosis
Jul 16, 2023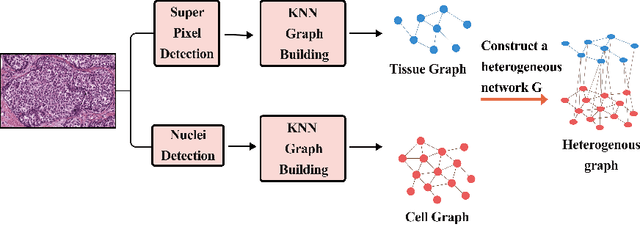
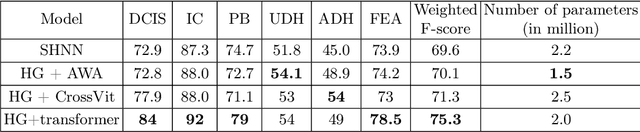
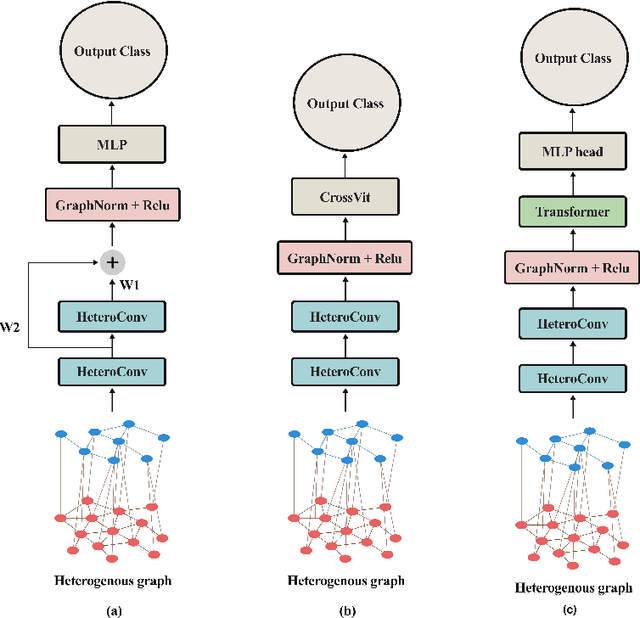
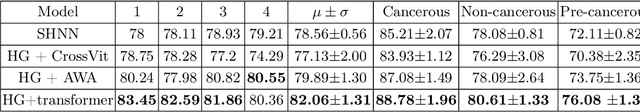
Abstract:The heterogeneity of breast cancer presents considerable challenges for its early detection, prognosis, and treatment selection. Convolutional neural networks often neglect the spatial relationships within histopathological images, which can limit their accuracy. Graph neural networks (GNNs) offer a promising solution by coding the spatial relationships within images. Prior studies have investigated the modeling of histopathological images as cell and tissue graphs, but they have not fully tapped into the potential of extracting interrelationships between these biological entities. In this paper, we present a novel approach using a heterogeneous GNN that captures the spatial and hierarchical relations between cell and tissue graphs to enhance the extraction of useful information from histopathological images. We also compare the performance of a cross-attention-based network and a transformer architecture for modeling the intricate relationships within tissue and cell graphs. Our model demonstrates superior efficiency in terms of parameter count and achieves higher accuracy compared to the transformer-based state-of-the-art approach on three publicly available breast cancer datasets -- BRIGHT, BreakHis, and BACH.
WaveMixSR: A Resource-efficient Neural Network for Image Super-resolution
Jul 01, 2023



Abstract:Image super-resolution research recently been dominated by transformer models which need higher computational resources than CNNs due to the quadratic complexity of self-attention. We propose a new neural network -- WaveMixSR -- for image super-resolution based on WaveMix architecture which uses a 2D-discrete wavelet transform for spatial token-mixing. Unlike transformer-based models, WaveMixSR does not unroll the image as a sequence of pixels/patches. It uses the inductive bias of convolutions along with the lossless token-mixing property of wavelet transform to achieve higher performance while requiring fewer resources and training data. We compare the performance of our network with other state-of-the-art methods for image super-resolution. Our experiments show that WaveMixSR achieves competitive performance in all datasets and reaches state-of-the-art performance in the BSD100 dataset on multiple super-resolution tasks. Our model is able to achieve this performance using less training data and computational resources while maintaining high parameter efficiency compared to current state-of-the-art models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge