Varsha S
Robust Semi-Supervised Learning for Histopathology Images through Self-Supervision Guided Out-of-Distribution Scoring
Mar 17, 2023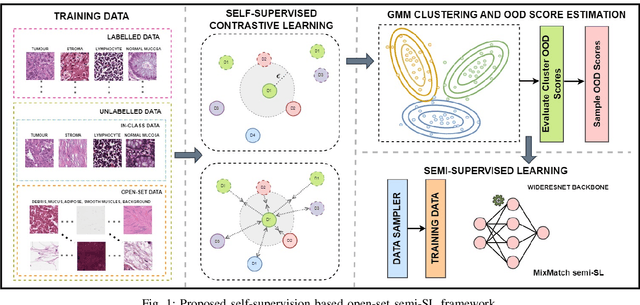
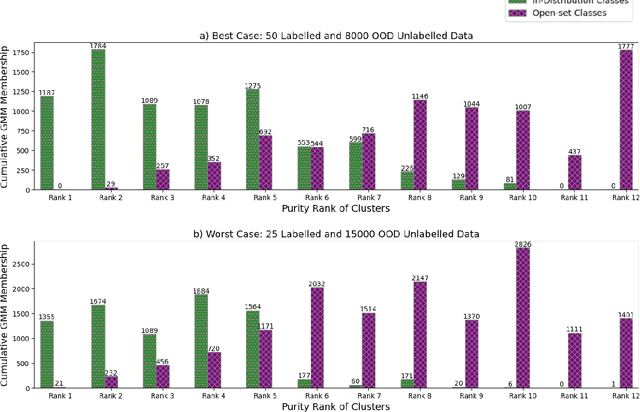
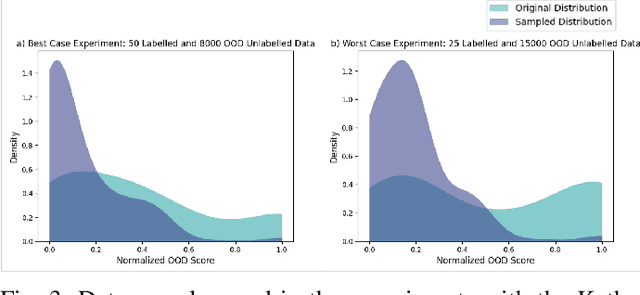
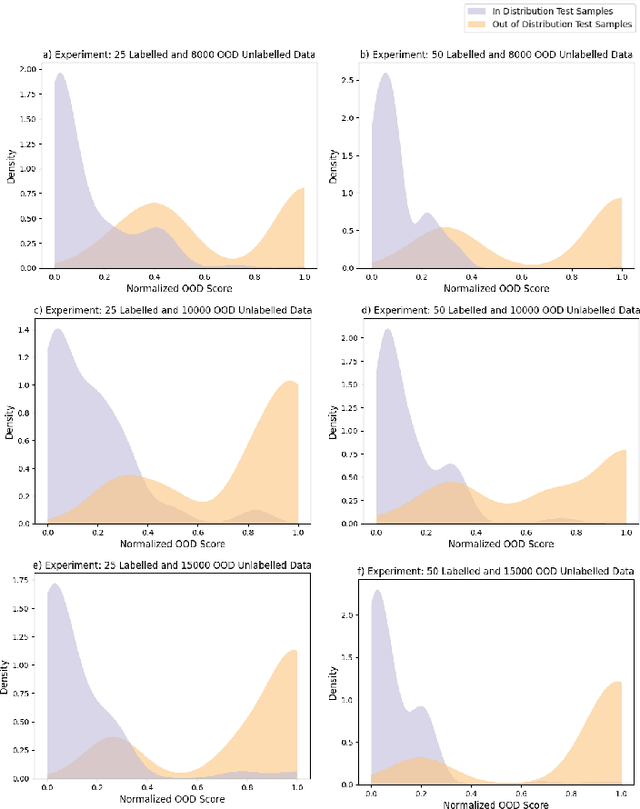
Abstract:Semi-supervised learning (semi-SL) is a promising alternative to supervised learning for medical image analysis when obtaining good quality supervision for medical imaging is difficult. However, semi-SL assumes that the underlying distribution of unaudited data matches that of the few labeled samples, which is often violated in practical settings, particularly in medical images. The presence of out-of-distribution (OOD) samples in the unlabeled training pool of semi-SL is inevitable and can reduce the efficiency of the algorithm. Common preprocessing methods to filter out outlier samples may not be suitable for medical images that involve a wide range of anatomical structures and rare morphologies. In this paper, we propose a novel pipeline for addressing open-set supervised learning challenges in digital histology images. Our pipeline efficiently estimates an OOD score for each unlabelled data point based on self-supervised learning to calibrate the knowledge needed for a subsequent semi-SL framework. The outlier score derived from the OOD detector is used to modulate sample selection for the subsequent semi-SL stage, ensuring that samples conforming to the distribution of the few labeled samples are more frequently exposed to the subsequent semi-SL framework. Our framework is compatible with any semi-SL framework, and we base our experiments on the popular Mixmatch semi-SL framework. We conduct extensive studies on two digital pathology datasets, Kather colorectal histology dataset and a dataset derived from TCGA-BRCA whole slide images, and establish the effectiveness of our method by comparing with popular methods and frameworks in semi-SL algorithms through various experiments.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge