Yaman Dang
Switching Loss for Generalized Nucleus Detection in Histopathology
Aug 09, 2020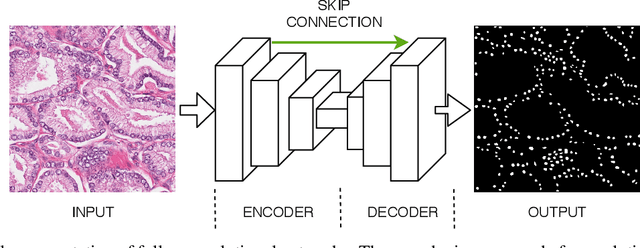
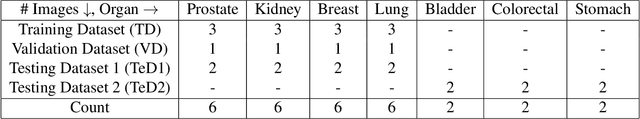
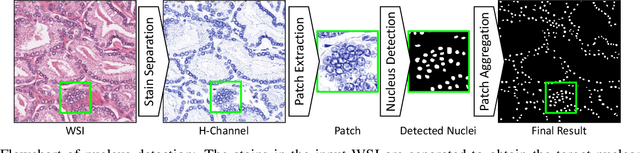
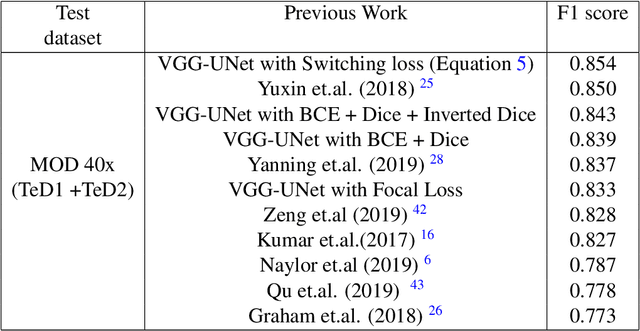
Abstract:The accuracy of deep learning methods for two foundational tasks in medical image analysis -- detection and segmentation -- can suffer from class imbalance. We propose a `switching loss' function that adaptively shifts the emphasis between foreground and background classes. While the existing loss functions to address this problem were motivated by the classification task, the switching loss is based on Dice loss, which is better suited for segmentation and detection. Furthermore, to get the most out the training samples, we adapt the loss with each mini-batch, unlike previous proposals that adapt once for the entire training set. A nucleus detector trained using the proposed loss function on a source dataset outperformed those trained using cross-entropy, Dice, or focal losses. Remarkably, without retraining on target datasets, our pre-trained nucleus detector also outperformed existing nucleus detectors that were trained on at least some of the images from the target datasets. To establish a broad utility of the proposed loss, we also confirmed that it led to more accurate ventricle segmentation in MRI as compared to the other loss functions. Our GPU-enabled pre-trained nucleus detection software is also ready to process whole slide images right out-of-the-box and is usably fast.
Pixel-wise Segmentation of Right Ventricle of Heart
Aug 21, 2019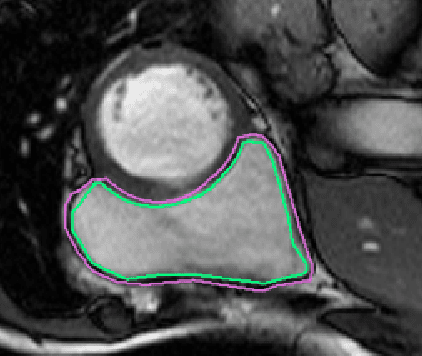
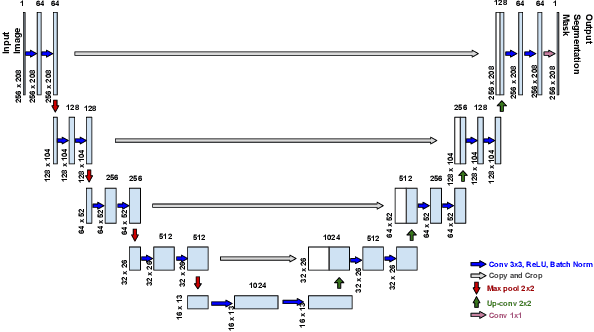
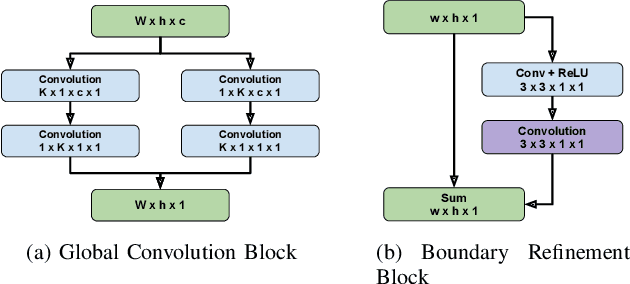
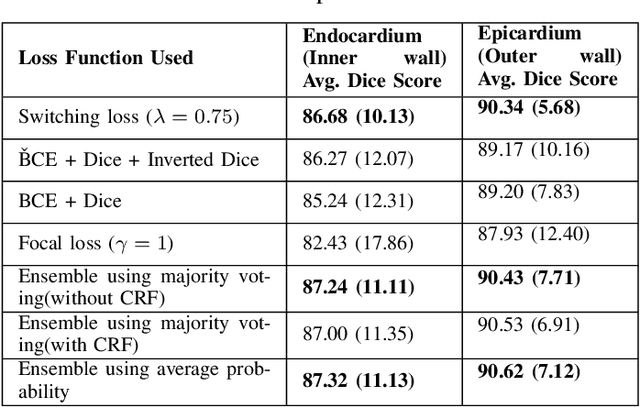
Abstract:One of the first steps in the diagnosis of most cardiac diseases, such as pulmonary hypertension, coronary heart disease is the segmentation of ventricles from cardiac magnetic resonance (MRI) images. Manual segmentation of the right ventricle requires diligence and time, while its automated segmentation is challenging due to shape variations and illdefined borders. We propose a deep learning based method for the accurate segmentation of right ventricle, which does not require post-processing and yet it achieves the state-of-the-art performance of 0.86 Dice coefficient and 6.73 mm Hausdorff distance on RVSC-MICCAI 2012 dataset. We use a novel adaptive cost function to counter extreme class-imbalance in the dataset. We present a comprehensive comparative study of loss functions, architectures, and ensembling techniques to build a principled approach for biomedical segmentation tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge