Shrey Gadiya
Histographs: Graphs in Histopathology
Aug 14, 2019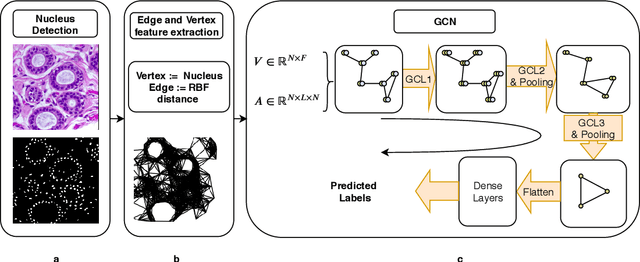
Abstract:Spatial arrangement of cells of various types, such as tumor infiltrating lymphocytes and the advancing edge of a tumor, are important features for detecting and characterizing cancers. However, convolutional neural networks (CNNs) do not explicitly extract intricate features of the spatial arrangements of the cells from histopathology images. In this work, we propose to classify cancers using graph convolutional networks (GCNs) by modeling a tissue section as a multi-attributed spatial graph of its constituent cells. Cells are detected using their nuclei in H&E stained tissue image, and each cell's appearance is captured as a multi-attributed high-dimensional vertex feature. The spatial relations between neighboring cells are captured as edge features based on their distances in a graph. We demonstrate the utility of this approach by obtaining classification accuracy that is competitive with CNNs, specifically, Inception-v3, on two tasks-cancerous versus non-cancerous and in situ versus invasive-on the BACH breast cancer dataset.
Some New Layer Architectures for Graph CNN
Oct 31, 2018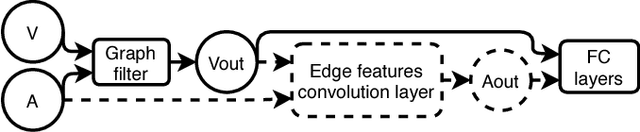



Abstract:While convolutional neural networks (CNNs) have recently made great strides in supervised classification of data structured on a grid (e.g. images composed of pixel grids), in several interesting datasets, the relations between features can be better represented as a general graph instead of a regular grid. Although recent algorithms that adapt CNNs to graphs have shown promising results, they mostly neglect learning explicit operations for edge features while focusing on vertex features alone. We propose new formulations for convolutional, pooling, and fully connected layers for neural networks that make more comprehensive use of the information available in multi-dimensional graphs. Using these layers led to an improvement in classification accuracy over the state-of-the-art methods on benchmark graph datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge