Fast GPU-Enabled Color Normalization for Digital Pathology
Paper and Code
Jan 10, 2019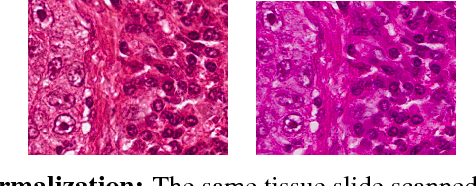
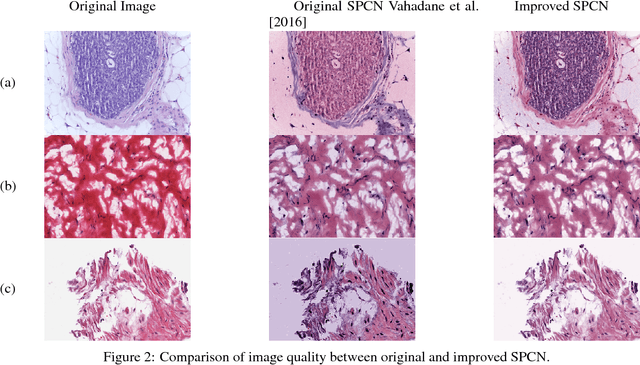

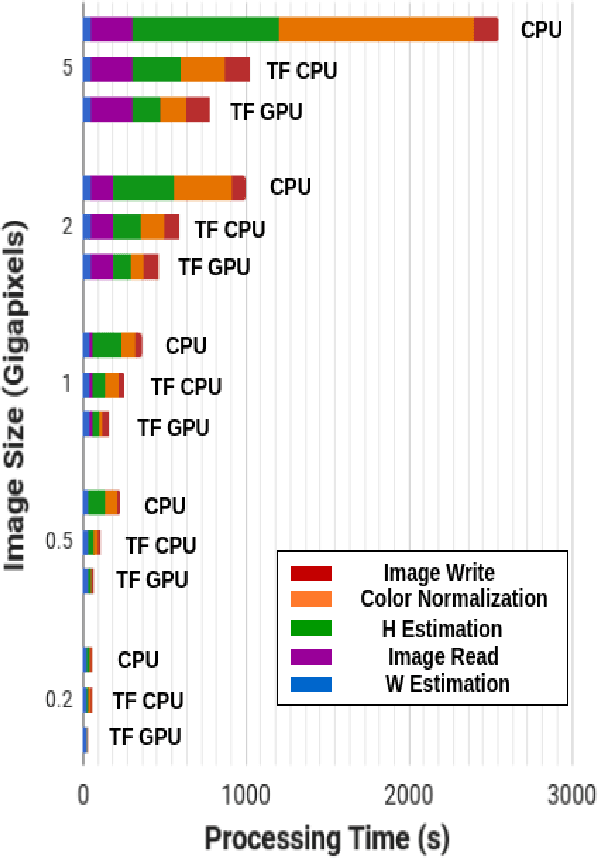
Normalizing unwanted color variations due to differences in staining processes and scanner responses has been shown to aid machine learning in computational pathology. Of the several popular techniques for color normalization, structure preserving color normalization (SPCN) is well-motivated, convincingly tested, and published with its code base. However, SPCN makes occasional errors in color basis estimation leading to artifacts such as swapping the color basis vectors between stains or giving a colored tinge to the background with no tissue. We made several algorithmic improvements to remove these artifacts. Additionally, the original SPCN code is not readily usable on gigapixel whole slide images (WSIs) due to long run times, use of proprietary software platform and libraries, and its inability to automatically handle WSIs. We completely rewrote the software such that it can automatically handle images of any size in popular WSI formats. Our software utilizes GPU-acceleration and open-source libraries that are becoming ubiquitous with the advent of deep learning. We also made several other small improvements and achieved a multifold overall speedup on gigapixel images. Our algorithm and software is usable right out-of-the-box by the computational pathology community.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge