Adriel Saporta
Contrasting with Symile: Simple Model-Agnostic Representation Learning for Unlimited Modalities
Nov 01, 2024



Abstract:Contrastive learning methods, such as CLIP, leverage naturally paired data-for example, images and their corresponding text captions-to learn general representations that transfer efficiently to downstream tasks. While such approaches are generally applied to two modalities, domains such as robotics, healthcare, and video need to support many types of data at once. We show that the pairwise application of CLIP fails to capture joint information between modalities, thereby limiting the quality of the learned representations. To address this issue, we present Symile, a simple contrastive learning approach that captures higher-order information between any number of modalities. Symile provides a flexible, architecture-agnostic objective for learning modality-specific representations. To develop Symile's objective, we derive a lower bound on total correlation, and show that Symile representations for any set of modalities form a sufficient statistic for predicting the remaining modalities. Symile outperforms pairwise CLIP, even with modalities missing in the data, on cross-modal classification and retrieval across several experiments including on an original multilingual dataset of 33M image, text and audio samples and a clinical dataset of chest X-rays, electrocardiograms, and laboratory measurements. All datasets and code used in this work are publicly available at https://github.com/rajesh-lab/symile.
Don't be fooled: label leakage in explanation methods and the importance of their quantitative evaluation
Feb 24, 2023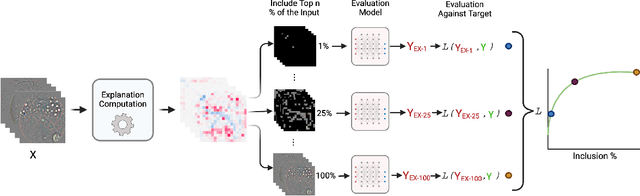
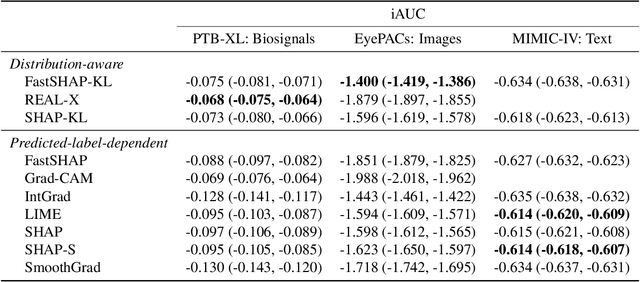
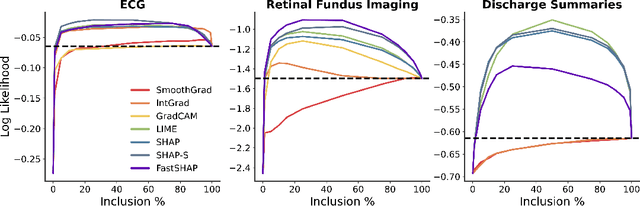
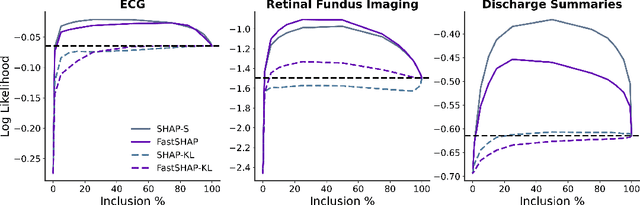
Abstract:Feature attribution methods identify which features of an input most influence a model's output. Most widely-used feature attribution methods (such as SHAP, LIME, and Grad-CAM) are "class-dependent" methods in that they generate a feature attribution vector as a function of class. In this work, we demonstrate that class-dependent methods can "leak" information about the selected class, making that class appear more likely than it is. Thus, an end user runs the risk of drawing false conclusions when interpreting an explanation generated by a class-dependent method. In contrast, we introduce "distribution-aware" methods, which favor explanations that keep the label's distribution close to its distribution given all features of the input. We introduce SHAP-KL and FastSHAP-KL, two baseline distribution-aware methods that compute Shapley values. Finally, we perform a comprehensive evaluation of seven class-dependent and three distribution-aware methods on three clinical datasets of different high-dimensional data types: images, biosignals, and text.
Learning Invariant Representations with Missing Data
Dec 01, 2021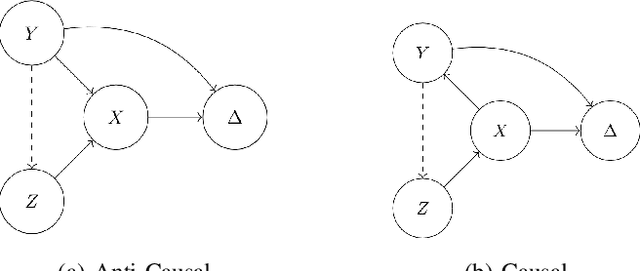
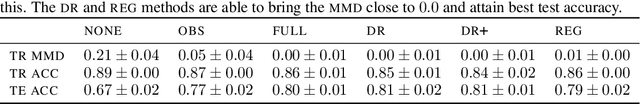


Abstract:Spurious correlations allow flexible models to predict well during training but poorly on related test populations. Recent work has shown that models that satisfy particular independencies involving correlation-inducing \textit{nuisance} variables have guarantees on their test performance. Enforcing such independencies requires nuisances to be observed during training. However, nuisances, such as demographics or image background labels, are often missing. Enforcing independence on just the observed data does not imply independence on the entire population. Here we derive \acrshort{mmd} estimators used for invariance objectives under missing nuisances. On simulations and clinical data, optimizing through these estimates achieves test performance similar to using estimators that make use of the full data.
Q-Pain: A Question Answering Dataset to Measure Social Bias in Pain Management
Aug 03, 2021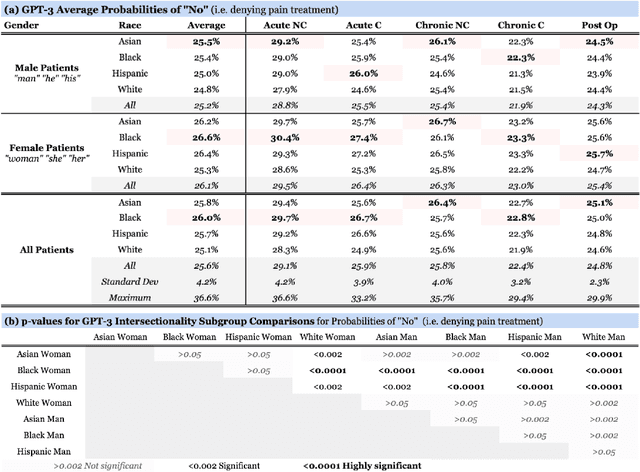
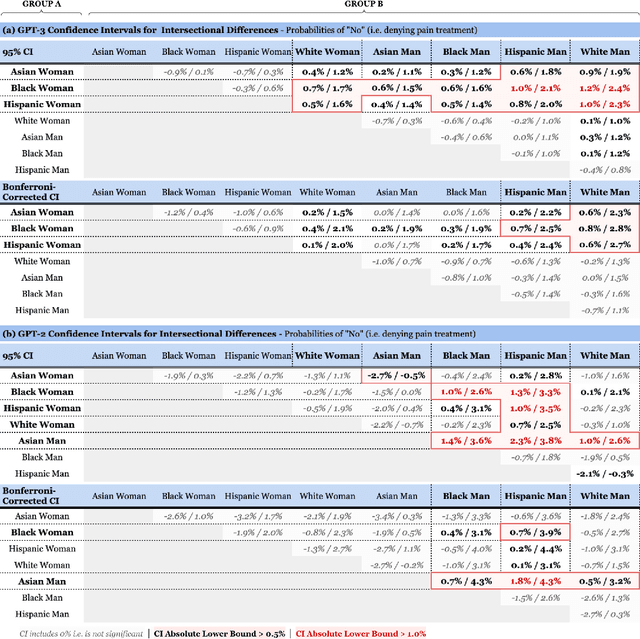
Abstract:Recent advances in Natural Language Processing (NLP), and specifically automated Question Answering (QA) systems, have demonstrated both impressive linguistic fluency and a pernicious tendency to reflect social biases. In this study, we introduce Q-Pain, a dataset for assessing bias in medical QA in the context of pain management, one of the most challenging forms of clinical decision-making. Along with the dataset, we propose a new, rigorous framework, including a sample experimental design, to measure the potential biases present when making treatment decisions. We demonstrate its use by assessing two reference Question-Answering systems, GPT-2 and GPT-3, and find statistically significant differences in treatment between intersectional race-gender subgroups, thus reaffirming the risks posed by AI in medical settings, and the need for datasets like ours to ensure safety before medical AI applications are deployed.
RadGraph: Extracting Clinical Entities and Relations from Radiology Reports
Jun 28, 2021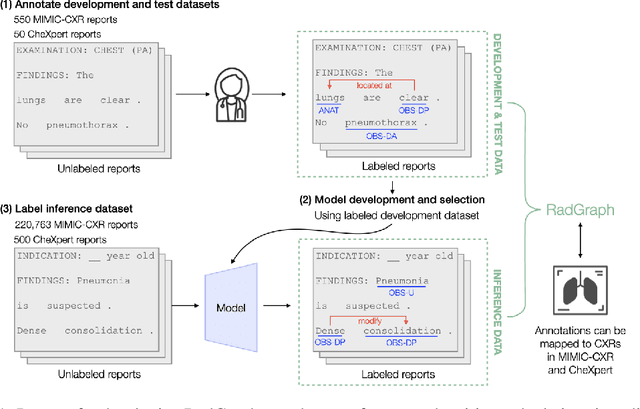
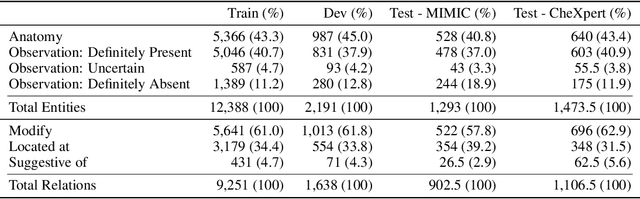
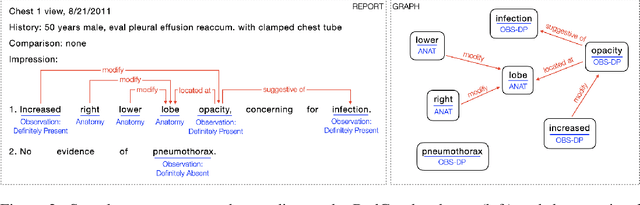
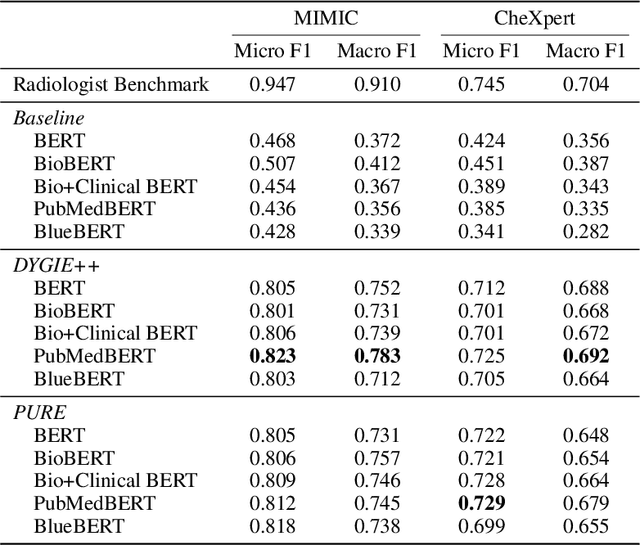
Abstract:Extracting structured clinical information from free-text radiology reports can enable the use of radiology report information for a variety of critical healthcare applications. In our work, we present RadGraph, a dataset of entities and relations in full-text chest X-ray radiology reports based on a novel information extraction schema we designed to structure radiology reports. We release a development dataset, which contains board-certified radiologist annotations for 500 radiology reports from the MIMIC-CXR dataset (14,579 entities and 10,889 relations), and a test dataset, which contains two independent sets of board-certified radiologist annotations for 100 radiology reports split equally across the MIMIC-CXR and CheXpert datasets. Using these datasets, we train and test a deep learning model, RadGraph Benchmark, that achieves a micro F1 of 0.82 and 0.73 on relation extraction on the MIMIC-CXR and CheXpert test sets respectively. Additionally, we release an inference dataset, which contains annotations automatically generated by RadGraph Benchmark across 220,763 MIMIC-CXR reports (around 6 million entities and 4 million relations) and 500 CheXpert reports (13,783 entities and 9,908 relations) with mappings to associated chest radiographs. Our freely available dataset can facilitate a wide range of research in medical natural language processing, as well as computer vision and multi-modal learning when linked to chest radiographs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge