Andrew C. Miller
Hybrid Modeling of Photoplethysmography for Non-invasive Monitoring of Cardiovascular Parameters
Nov 18, 2025Abstract:Continuous cardiovascular monitoring can play a key role in precision health. However, some fundamental cardiac biomarkers of interest, including stroke volume and cardiac output, require invasive measurements, e.g., arterial pressure waveforms (APW). As a non-invasive alternative, photoplethysmography (PPG) measurements are routinely collected in hospital settings. Unfortunately, the prediction of key cardiac biomarkers from PPG instead of APW remains an open challenge, further complicated by the scarcity of annotated PPG measurements. As a solution, we propose a hybrid approach that uses hemodynamic simulations and unlabeled clinical data to estimate cardiovascular biomarkers directly from PPG signals. Our hybrid model combines a conditional variational autoencoder trained on paired PPG-APW data with a conditional density estimator of cardiac biomarkers trained on labeled simulated APW segments. As a key result, our experiments demonstrate that the proposed approach can detect fluctuations of cardiac output and stroke volume and outperform a supervised baseline in monitoring temporal changes in these biomarkers.
Inferring Optical Tissue Properties from Photoplethysmography using Hybrid Amortized Inference
Oct 02, 2025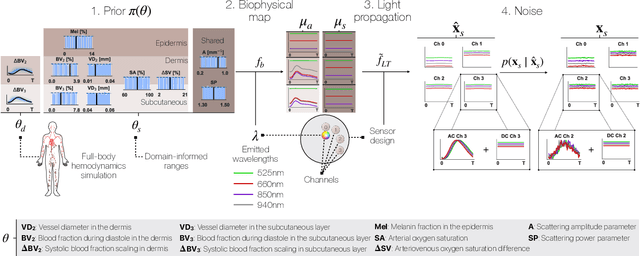
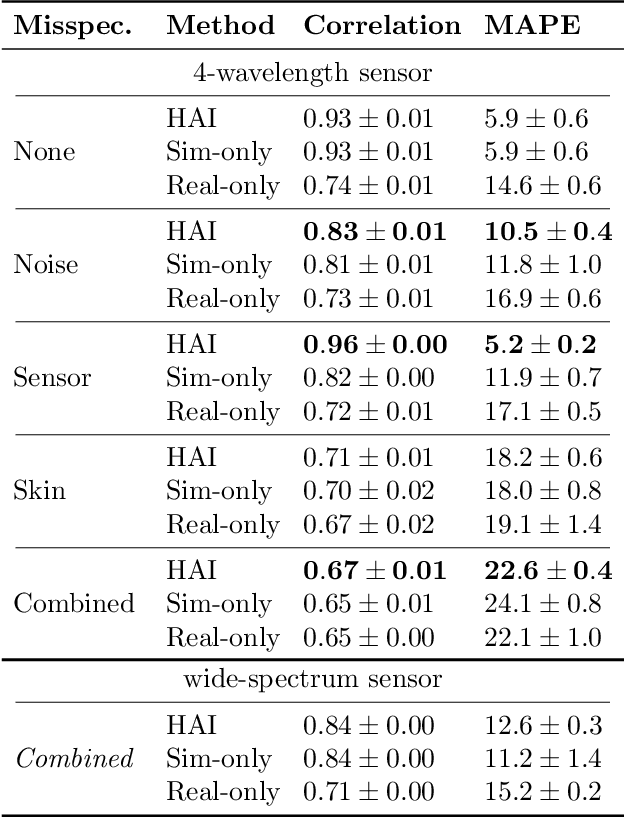
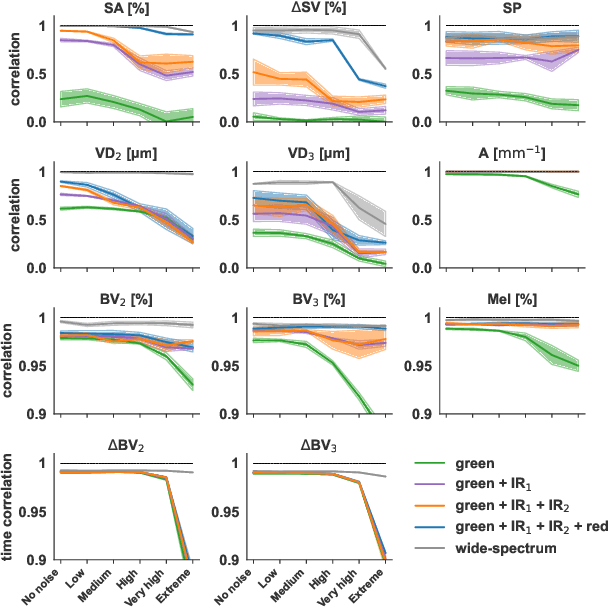

Abstract:Smart wearables enable continuous tracking of established biomarkers such as heart rate, heart rate variability, and blood oxygen saturation via photoplethysmography (PPG). Beyond these metrics, PPG waveforms contain richer physiological information, as recent deep learning (DL) studies demonstrate. However, DL models often rely on features with unclear physiological meaning, creating a tension between predictive power, clinical interpretability, and sensor design. We address this gap by introducing PPGen, a biophysical model that relates PPG signals to interpretable physiological and optical parameters. Building on PPGen, we propose hybrid amortized inference (HAI), enabling fast, robust, and scalable estimation of relevant physiological parameters from PPG signals while correcting for model misspecification. In extensive in-silico experiments, we show that HAI can accurately infer physiological parameters under diverse noise and sensor conditions. Our results illustrate a path toward PPG models that retain the fidelity needed for DL-based features while supporting clinical interpretation and informed hardware design.
Wearable Accelerometer Foundation Models for Health via Knowledge Distillation
Dec 15, 2024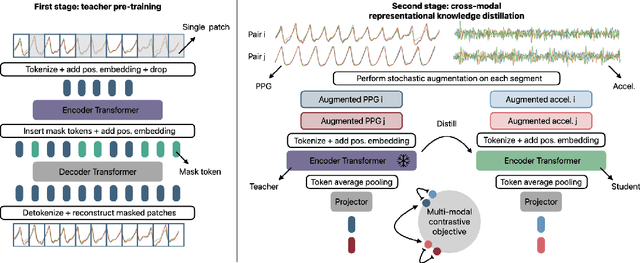

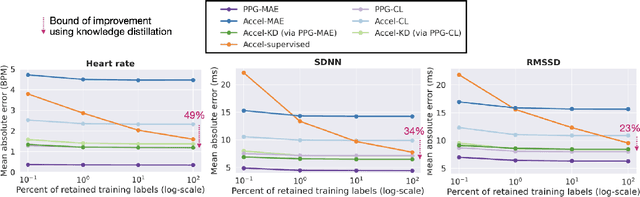
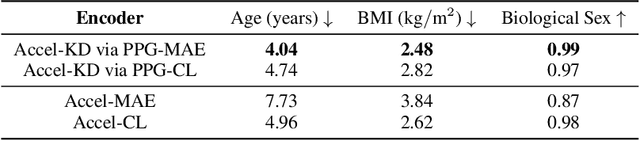
Abstract:Modern wearable devices can conveniently and continuously record various biosignals in the many different environments of daily living, ultimately enabling a rich view of individual health. However, not all biosignals are the same: high-fidelity measurements, such as photoplethysmography (PPG), contain more physiological information, but require optical sensors with a high power footprint. In a resource-constrained setting, such biosignals may be unavailable. Alternatively, a lower-fidelity biosignal, such as accelerometry that captures minute cardiovascular information during low-motion periods, has a significantly smaller power footprint and is available in almost any wearable device. Here, we demonstrate that we can distill representational knowledge across biosignals, i.e., from PPG to accelerometry, using 20 million minutes of unlabeled data, collected from ~172K participants in the Apple Heart and Movement Study under informed consent. We first pre-train PPG encoders via self-supervised learning, and then distill their representational knowledge to accelerometry encoders. We demonstrate strong cross-modal alignment on unseen data, e.g., 99.2% top-1 accuracy for retrieving PPG embeddings from accelerometry embeddings. We show that distilled accelerometry encoders have significantly more informative representations compared to self-supervised or supervised encoders trained directly on accelerometry data, observed by at least 23%-49% improved performance for predicting heart rate and heart rate variability. We also show that distilled accelerometry encoders are readily predictive of a wide array of downstream health targets, i.e., they are generalist foundation models. We believe accelerometry foundation models for health may unlock new opportunities for developing digital biomarkers from any wearable device, and help individuals track their health more frequently and conveniently.
Large-scale Training of Foundation Models for Wearable Biosignals
Dec 08, 2023



Abstract:Tracking biosignals is crucial for monitoring wellness and preempting the development of severe medical conditions. Today, wearable devices can conveniently record various biosignals, creating the opportunity to monitor health status without disruption to one's daily routine. Despite widespread use of wearable devices and existing digital biomarkers, the absence of curated data with annotated medical labels hinders the development of new biomarkers to measure common health conditions. In fact, medical datasets are usually small in comparison to other domains, which is an obstacle for developing neural network models for biosignals. To address this challenge, we have employed self-supervised learning using the unlabeled sensor data collected under informed consent from the large longitudinal Apple Heart and Movement Study (AHMS) to train foundation models for two common biosignals: photoplethysmography (PPG) and electrocardiogram (ECG) recorded on Apple Watch. We curated PPG and ECG datasets from AHMS that include data from ~141K participants spanning ~3 years. Our self-supervised learning framework includes participant level positive pair selection, stochastic augmentation module and a regularized contrastive loss optimized with momentum training, and generalizes well to both PPG and ECG modalities. We show that the pre-trained foundation models readily encode information regarding participants' demographics and health conditions. To the best of our knowledge, this is the first study that builds foundation models using large-scale PPG and ECG data collected via wearable consumer devices $\unicode{x2013}$ prior works have commonly used smaller-size datasets collected in clinical and experimental settings. We believe PPG and ECG foundation models can enhance future wearable devices by reducing the reliance on labeled data and hold the potential to help the users improve their health.
Label Shift Estimators for Non-Ignorable Missing Data
Oct 27, 2023



Abstract:We consider the problem of estimating the mean of a random variable Y subject to non-ignorable missingness, i.e., where the missingness mechanism depends on Y . We connect the auxiliary proxy variable framework for non-ignorable missingness (West and Little, 2013) to the label shift setting (Saerens et al., 2002). Exploiting this connection, we construct an estimator for non-ignorable missing data that uses high-dimensional covariates (or proxies) without the need for a generative model. In synthetic and semi-synthetic experiments, we study the behavior of the proposed estimator, comparing it to commonly used ignorable estimators in both well-specified and misspecified settings. Additionally, we develop a score to assess how consistent the data are with the label shift assumption. We use our approach to estimate disease prevalence using a large health survey, comparing ignorable and non-ignorable approaches. We show that failing to account for non-ignorable missingness can have profound consequences on conclusions drawn from non-representative samples.
Simulation-based Inference for Cardiovascular Models
Jul 29, 2023Abstract:Over the past decades, hemodynamics simulators have steadily evolved and have become tools of choice for studying cardiovascular systems in-silico. While such tools are routinely used to simulate whole-body hemodynamics from physiological parameters, solving the corresponding inverse problem of mapping waveforms back to plausible physiological parameters remains both promising and challenging. Motivated by advances in simulation-based inference (SBI), we cast this inverse problem as statistical inference. In contrast to alternative approaches, SBI provides \textit{posterior distributions} for the parameters of interest, providing a \textit{multi-dimensional} representation of uncertainty for \textit{individual} measurements. We showcase this ability by performing an in-silico uncertainty analysis of five biomarkers of clinical interest comparing several measurement modalities. Beyond the corroboration of known facts, such as the feasibility of estimating heart rate, our study highlights the potential of estimating new biomarkers from standard-of-care measurements. SBI reveals practically relevant findings that cannot be captured by standard sensitivity analyses, such as the existence of sub-populations for which parameter estimation exhibits distinct uncertainty regimes. Finally, we study the gap between in-vivo and in-silico with the MIMIC-III waveform database and critically discuss how cardiovascular simulations can inform real-world data analysis.
Learning Invariant Representations with Missing Data
Dec 01, 2021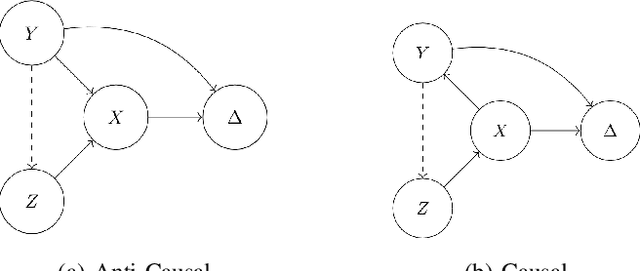
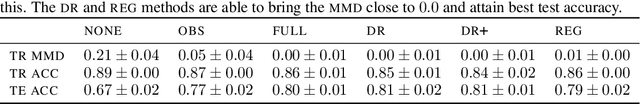


Abstract:Spurious correlations allow flexible models to predict well during training but poorly on related test populations. Recent work has shown that models that satisfy particular independencies involving correlation-inducing \textit{nuisance} variables have guarantees on their test performance. Enforcing such independencies requires nuisances to be observed during training. However, nuisances, such as demographics or image background labels, are often missing. Enforcing independence on just the observed data does not imply independence on the entire population. Here we derive \acrshort{mmd} estimators used for invariance objectives under missing nuisances. On simulations and clinical data, optimizing through these estimates achieves test performance similar to using estimators that make use of the full data.
Model-based metrics: Sample-efficient estimates of predictive model subpopulation performance
Apr 25, 2021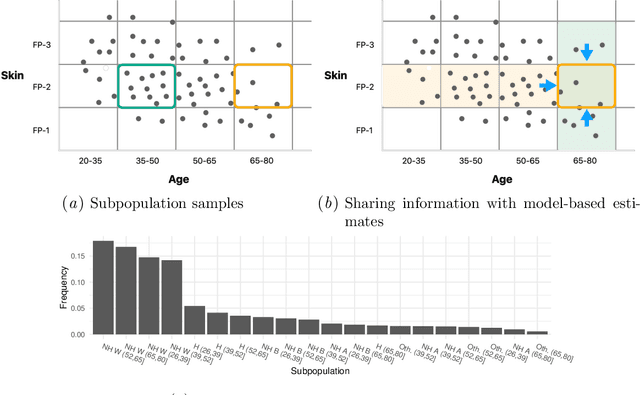
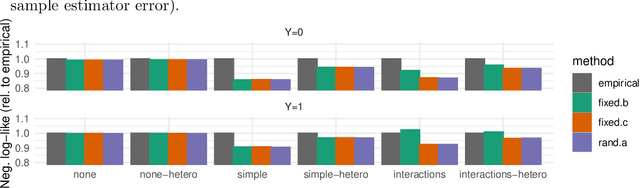
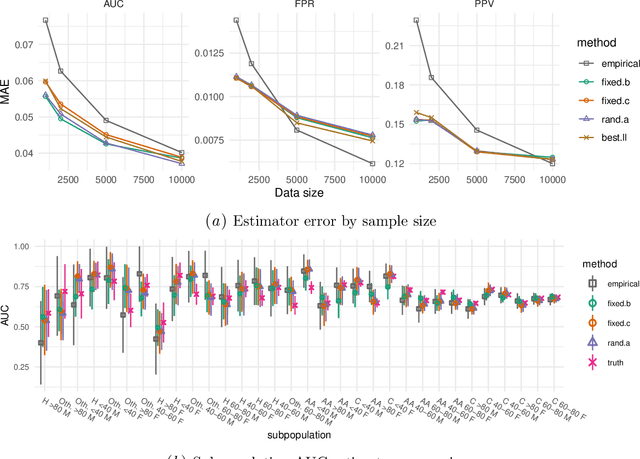
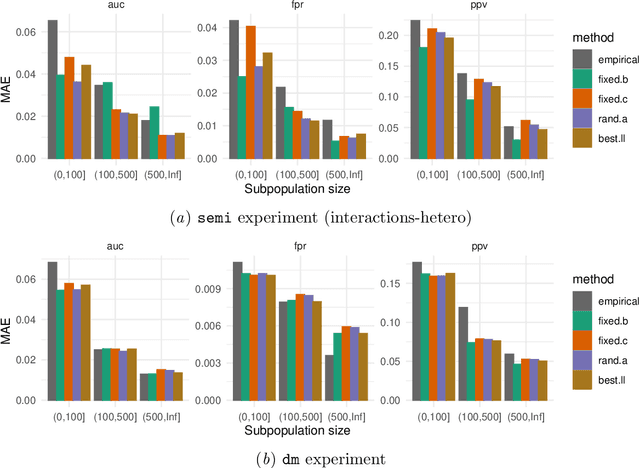
Abstract:Machine learning models $-$ now commonly developed to screen, diagnose, or predict health conditions $-$ are evaluated with a variety of performance metrics. An important first step in assessing the practical utility of a model is to evaluate its average performance over an entire population of interest. In many settings, it is also critical that the model makes good predictions within predefined subpopulations. For instance, showing that a model is fair or equitable requires evaluating the model's performance in different demographic subgroups. However, subpopulation performance metrics are typically computed using only data from that subgroup, resulting in higher variance estimates for smaller groups. We devise a procedure to measure subpopulation performance that can be more sample-efficient than the typical subsample estimates. We propose using an evaluation model $-$ a model that describes the conditional distribution of the predictive model score $-$ to form model-based metric (MBM) estimates. Our procedure incorporates model checking and validation, and we propose a computationally efficient approximation of the traditional nonparametric bootstrap to form confidence intervals. We evaluate MBMs on two main tasks: a semi-synthetic setting where ground truth metrics are available and a real-world hospital readmission prediction task. We find that MBMs consistently produce more accurate and lower variance estimates of model performance for small subpopulations.
Breiman's two cultures: You don't have to choose sides
Apr 25, 2021Abstract:Breiman's classic paper casts data analysis as a choice between two cultures: data modelers and algorithmic modelers. Stated broadly, data modelers use simple, interpretable models with well-understood theoretical properties to analyze data. Algorithmic modelers prioritize predictive accuracy and use more flexible function approximations to analyze data. This dichotomy overlooks a third set of models $-$ mechanistic models derived from scientific theories (e.g., ODE/SDE simulators). Mechanistic models encode application-specific scientific knowledge about the data. And while these categories represent extreme points in model space, modern computational and algorithmic tools enable us to interpolate between these points, producing flexible, interpretable, and scientifically-informed hybrids that can enjoy accurate and robust predictions, and resolve issues with data analysis that Breiman describes, such as the Rashomon effect and Occam's dilemma. Challenges still remain in finding an appropriate point in model space, with many choices on how to compose model components and the degree to which each component informs inferences.
Representing and Denoising Wearable ECG Recordings
Nov 30, 2020



Abstract:Modern wearable devices are embedded with a range of noninvasive biomarker sensors that hold promise for improving detection and treatment of disease. One such sensor is the single-lead electrocardiogram (ECG) which measures electrical signals in the heart. The benefits of the sheer volume of ECG measurements with rich longitudinal structure made possible by wearables come at the price of potentially noisier measurements compared to clinical ECGs, e.g., due to movement. In this work, we develop a statistical model to simulate a structured noise process in ECGs derived from a wearable sensor, design a beat-to-beat representation that is conducive for analyzing variation, and devise a factor analysis-based method to denoise the ECG. We study synthetic data generated using a realistic ECG simulator and a structured noise model. At varying levels of signal-to-noise, we quantitatively measure an upper bound on performance and compare estimates from linear and non-linear models. Finally, we apply our method to a set of ECGs collected by wearables in a mobile health study.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge