Salar Abbaspourazad
Wearable Accelerometer Foundation Models for Health via Knowledge Distillation
Dec 15, 2024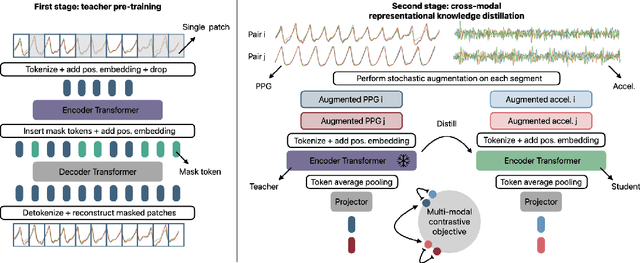

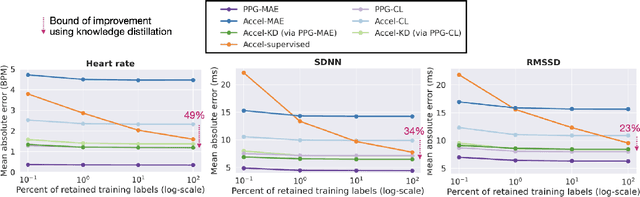
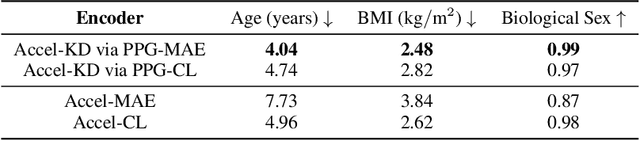
Abstract:Modern wearable devices can conveniently and continuously record various biosignals in the many different environments of daily living, ultimately enabling a rich view of individual health. However, not all biosignals are the same: high-fidelity measurements, such as photoplethysmography (PPG), contain more physiological information, but require optical sensors with a high power footprint. In a resource-constrained setting, such biosignals may be unavailable. Alternatively, a lower-fidelity biosignal, such as accelerometry that captures minute cardiovascular information during low-motion periods, has a significantly smaller power footprint and is available in almost any wearable device. Here, we demonstrate that we can distill representational knowledge across biosignals, i.e., from PPG to accelerometry, using 20 million minutes of unlabeled data, collected from ~172K participants in the Apple Heart and Movement Study under informed consent. We first pre-train PPG encoders via self-supervised learning, and then distill their representational knowledge to accelerometry encoders. We demonstrate strong cross-modal alignment on unseen data, e.g., 99.2% top-1 accuracy for retrieving PPG embeddings from accelerometry embeddings. We show that distilled accelerometry encoders have significantly more informative representations compared to self-supervised or supervised encoders trained directly on accelerometry data, observed by at least 23%-49% improved performance for predicting heart rate and heart rate variability. We also show that distilled accelerometry encoders are readily predictive of a wide array of downstream health targets, i.e., they are generalist foundation models. We believe accelerometry foundation models for health may unlock new opportunities for developing digital biomarkers from any wearable device, and help individuals track their health more frequently and conveniently.
Large-scale Training of Foundation Models for Wearable Biosignals
Dec 08, 2023



Abstract:Tracking biosignals is crucial for monitoring wellness and preempting the development of severe medical conditions. Today, wearable devices can conveniently record various biosignals, creating the opportunity to monitor health status without disruption to one's daily routine. Despite widespread use of wearable devices and existing digital biomarkers, the absence of curated data with annotated medical labels hinders the development of new biomarkers to measure common health conditions. In fact, medical datasets are usually small in comparison to other domains, which is an obstacle for developing neural network models for biosignals. To address this challenge, we have employed self-supervised learning using the unlabeled sensor data collected under informed consent from the large longitudinal Apple Heart and Movement Study (AHMS) to train foundation models for two common biosignals: photoplethysmography (PPG) and electrocardiogram (ECG) recorded on Apple Watch. We curated PPG and ECG datasets from AHMS that include data from ~141K participants spanning ~3 years. Our self-supervised learning framework includes participant level positive pair selection, stochastic augmentation module and a regularized contrastive loss optimized with momentum training, and generalizes well to both PPG and ECG modalities. We show that the pre-trained foundation models readily encode information regarding participants' demographics and health conditions. To the best of our knowledge, this is the first study that builds foundation models using large-scale PPG and ECG data collected via wearable consumer devices $\unicode{x2013}$ prior works have commonly used smaller-size datasets collected in clinical and experimental settings. We believe PPG and ECG foundation models can enhance future wearable devices by reducing the reliance on labeled data and hold the potential to help the users improve their health.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge