Joseph Futoma
Wearable Accelerometer Foundation Models for Health via Knowledge Distillation
Dec 15, 2024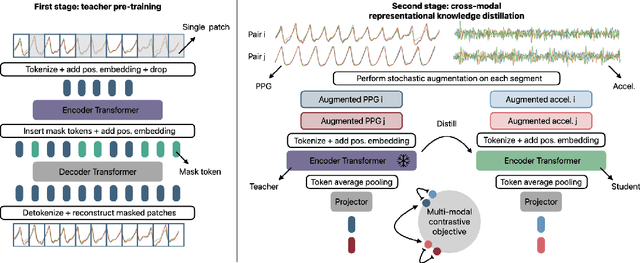

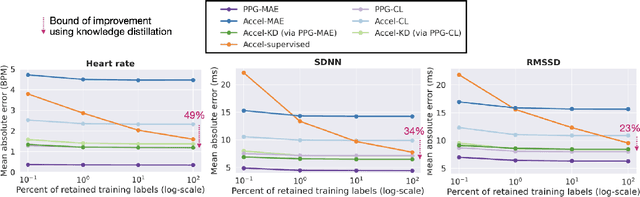
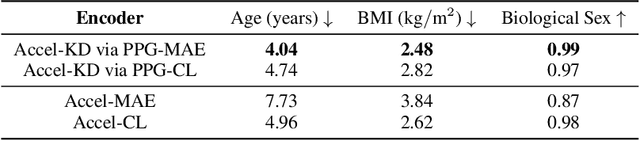
Abstract:Modern wearable devices can conveniently and continuously record various biosignals in the many different environments of daily living, ultimately enabling a rich view of individual health. However, not all biosignals are the same: high-fidelity measurements, such as photoplethysmography (PPG), contain more physiological information, but require optical sensors with a high power footprint. In a resource-constrained setting, such biosignals may be unavailable. Alternatively, a lower-fidelity biosignal, such as accelerometry that captures minute cardiovascular information during low-motion periods, has a significantly smaller power footprint and is available in almost any wearable device. Here, we demonstrate that we can distill representational knowledge across biosignals, i.e., from PPG to accelerometry, using 20 million minutes of unlabeled data, collected from ~172K participants in the Apple Heart and Movement Study under informed consent. We first pre-train PPG encoders via self-supervised learning, and then distill their representational knowledge to accelerometry encoders. We demonstrate strong cross-modal alignment on unseen data, e.g., 99.2% top-1 accuracy for retrieving PPG embeddings from accelerometry embeddings. We show that distilled accelerometry encoders have significantly more informative representations compared to self-supervised or supervised encoders trained directly on accelerometry data, observed by at least 23%-49% improved performance for predicting heart rate and heart rate variability. We also show that distilled accelerometry encoders are readily predictive of a wide array of downstream health targets, i.e., they are generalist foundation models. We believe accelerometry foundation models for health may unlock new opportunities for developing digital biomarkers from any wearable device, and help individuals track their health more frequently and conveniently.
Label Shift Estimators for Non-Ignorable Missing Data
Oct 27, 2023



Abstract:We consider the problem of estimating the mean of a random variable Y subject to non-ignorable missingness, i.e., where the missingness mechanism depends on Y . We connect the auxiliary proxy variable framework for non-ignorable missingness (West and Little, 2013) to the label shift setting (Saerens et al., 2002). Exploiting this connection, we construct an estimator for non-ignorable missing data that uses high-dimensional covariates (or proxies) without the need for a generative model. In synthetic and semi-synthetic experiments, we study the behavior of the proposed estimator, comparing it to commonly used ignorable estimators in both well-specified and misspecified settings. Additionally, we develop a score to assess how consistent the data are with the label shift assumption. We use our approach to estimate disease prevalence using a large health survey, comparing ignorable and non-ignorable approaches. We show that failing to account for non-ignorable missingness can have profound consequences on conclusions drawn from non-representative samples.
Model-based metrics: Sample-efficient estimates of predictive model subpopulation performance
Apr 25, 2021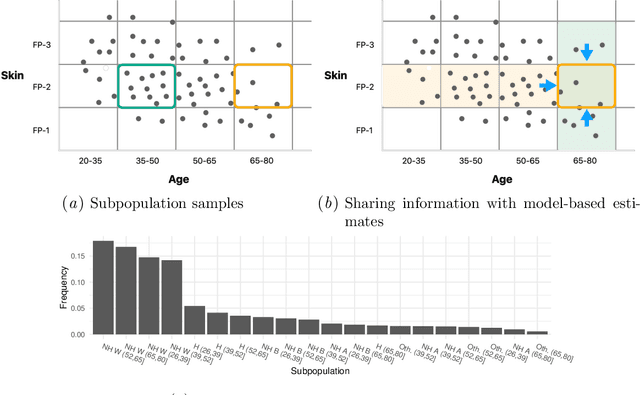
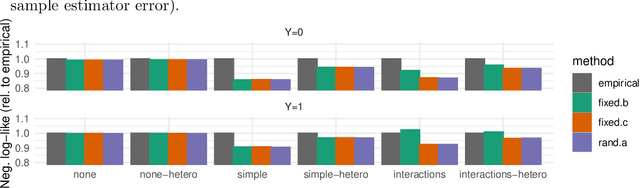
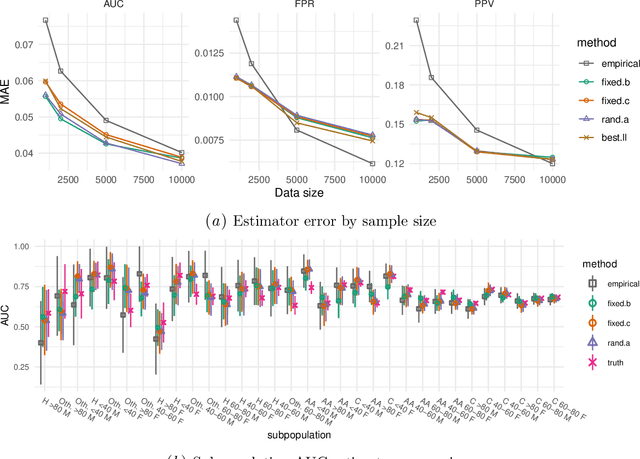
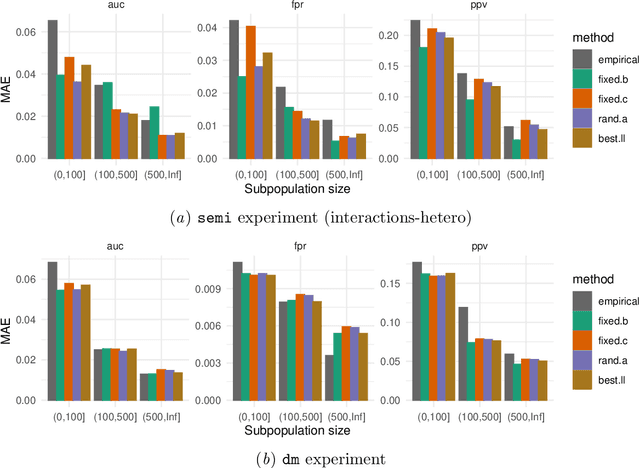
Abstract:Machine learning models $-$ now commonly developed to screen, diagnose, or predict health conditions $-$ are evaluated with a variety of performance metrics. An important first step in assessing the practical utility of a model is to evaluate its average performance over an entire population of interest. In many settings, it is also critical that the model makes good predictions within predefined subpopulations. For instance, showing that a model is fair or equitable requires evaluating the model's performance in different demographic subgroups. However, subpopulation performance metrics are typically computed using only data from that subgroup, resulting in higher variance estimates for smaller groups. We devise a procedure to measure subpopulation performance that can be more sample-efficient than the typical subsample estimates. We propose using an evaluation model $-$ a model that describes the conditional distribution of the predictive model score $-$ to form model-based metric (MBM) estimates. Our procedure incorporates model checking and validation, and we propose a computationally efficient approximation of the traditional nonparametric bootstrap to form confidence intervals. We evaluate MBMs on two main tasks: a semi-synthetic setting where ground truth metrics are available and a real-world hospital readmission prediction task. We find that MBMs consistently produce more accurate and lower variance estimates of model performance for small subpopulations.
Model-based Reinforcement Learning for Semi-Markov Decision Processes with Neural ODEs
Jun 29, 2020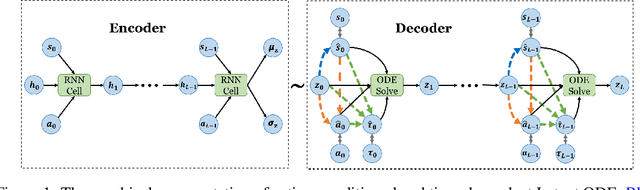
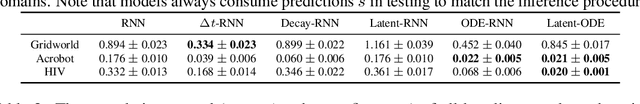
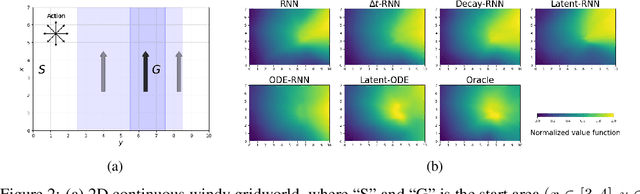
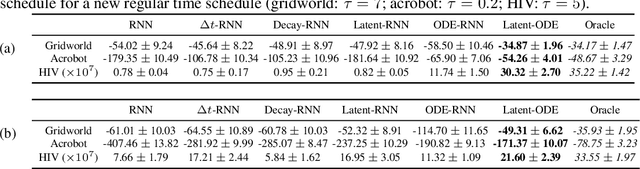
Abstract:We present two elegant solutions for modeling continuous-time dynamics, in a novel model-based reinforcement learning (RL) framework for semi-Markov decision processes (SMDPs), using neural ordinary differential equations (ODEs). Our models accurately characterize continuous-time dynamics and enable us to develop high-performing policies using a small amount of data. We also develop a model-based approach for optimizing time schedules to reduce interaction rates with the environment while maintaining the near-optimal performance, which is not possible for model-free methods. We experimentally demonstrate the efficacy of our methods across various continuous-time domains.
Interpretable Off-Policy Evaluation in Reinforcement Learning by Highlighting Influential Transitions
Feb 14, 2020
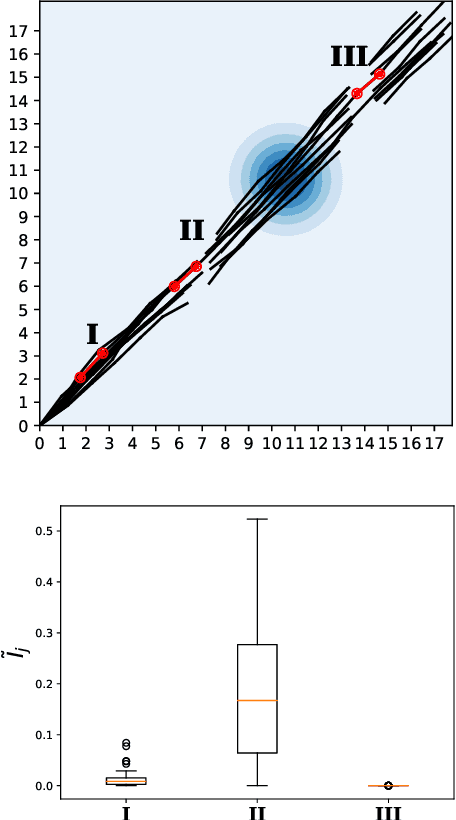
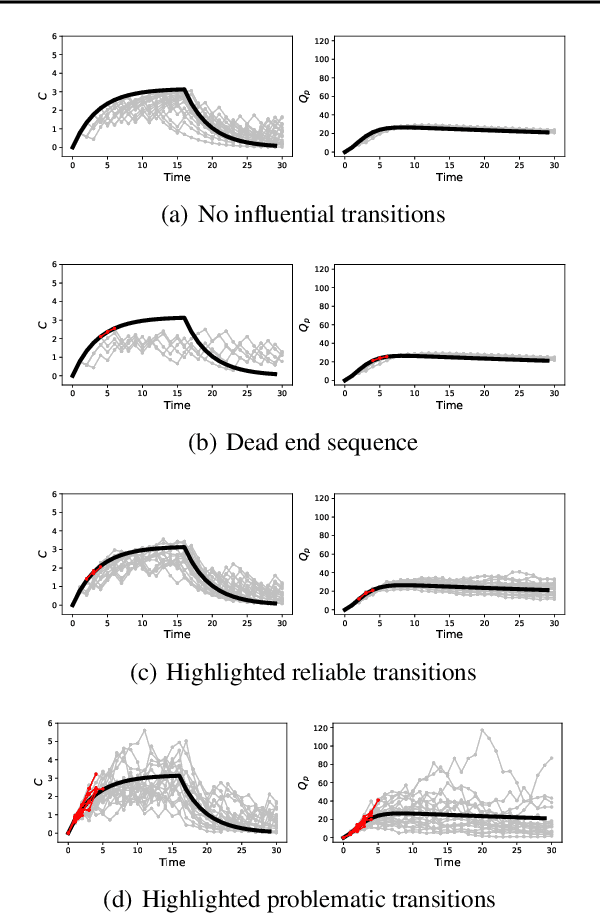
Abstract:Off-policy evaluation in reinforcement learning offers the chance of using observational data to improve future outcomes in domains such as healthcare and education, but safe deployment in high stakes settings requires ways of assessing its validity. Traditional measures such as confidence intervals may be insufficient due to noise, limited data and confounding. In this paper we develop a method that could serve as a hybrid human-AI system, to enable human experts to analyze the validity of policy evaluation estimates. This is accomplished by highlighting observations in the data whose removal will have a large effect on the OPE estimate, and formulating a set of rules for choosing which ones to present to domain experts for validation. We develop methods to compute exactly the influence functions for fitted Q-evaluation with two different function classes: kernel-based and linear least squares. Experiments on medical simulations and real-world intensive care unit data demonstrate that our method can be used to identify limitations in the evaluation process and make evaluation more robust.
POPCORN: Partially Observed Prediction COnstrained ReiNforcement Learning
Jan 13, 2020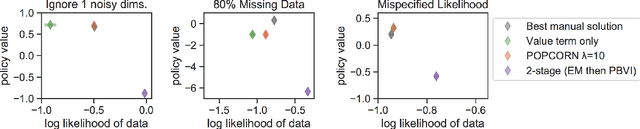
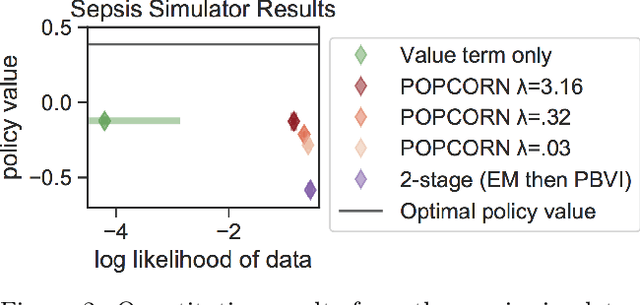
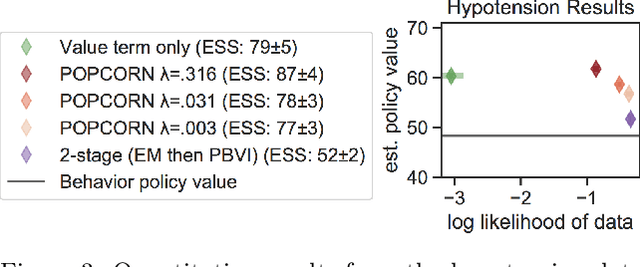
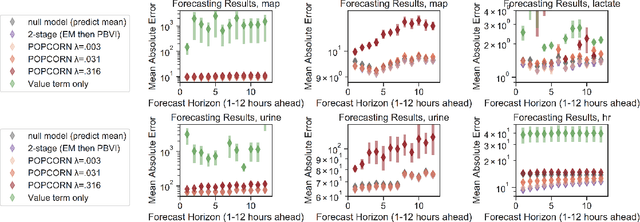
Abstract:Many medical decision-making settings can be framed as partially observed Markov decision processes (POMDPs). However, popular two-stage approaches that first learn a POMDP model and then solve it often fail because the model that best fits the data may not be the best model for planning. We introduce a new optimization objective that (a) produces both high-performing policies and high-quality generative models, even when some observations are irrelevant for planning, and (b) does so in the kinds of batch, off-policy settings common in medicine. We demonstrate our approach on synthetic examples and a real-world hypotension management task.
Identifying Distinct, Effective Treatments for Acute Hypotension with SODA-RL: Safely Optimized Diverse Accurate Reinforcement Learning
Jan 09, 2020

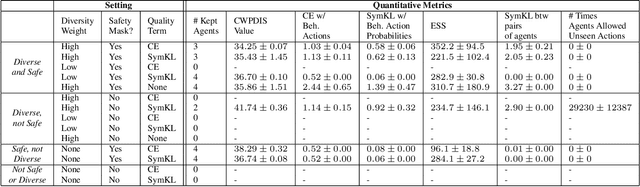
Abstract:Hypotension in critical care settings is a life-threatening emergency that must be recognized and treated early. While fluid bolus therapy and vasopressors are common treatments, it is often unclear which interventions to give, in what amounts, and for how long. Observational data in the form of electronic health records can provide a source for helping inform these choices from past events, but often it is not possible to identify a single best strategy from observational data alone. In such situations, we argue it is important to expose the collection of plausible options to a provider. To this end, we develop SODA-RL: Safely Optimized, Diverse, and Accurate Reinforcement Learning, to identify distinct treatment options that are supported in the data. We demonstrate SODA-RL on a cohort of 10,142 ICU stays where hypotension presented. Our learned policies perform comparably to the observed physician behaviors, while providing different, plausible alternatives for treatment decisions.
"The Human Body is a Black Box": Supporting Clinical Decision-Making with Deep Learning
Dec 07, 2019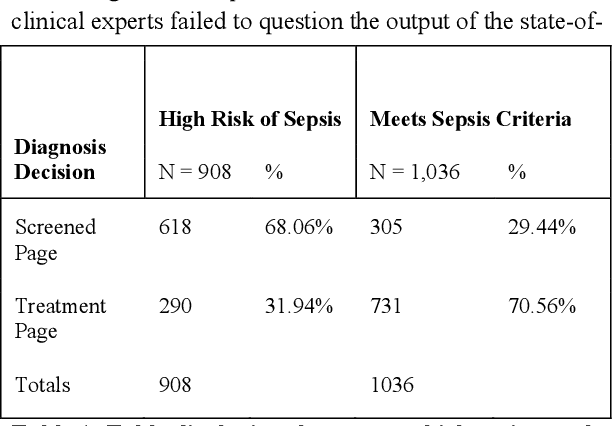
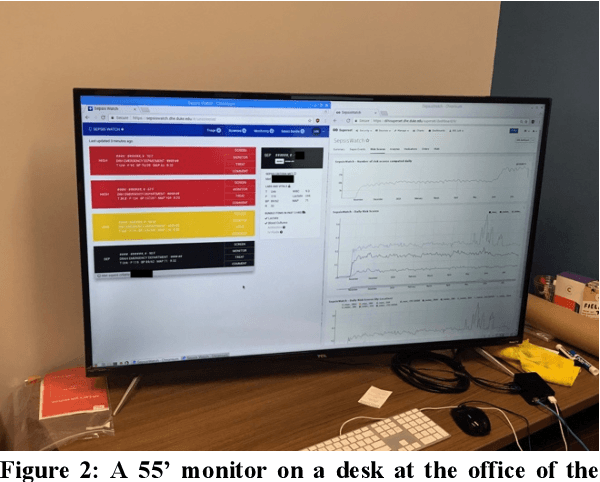
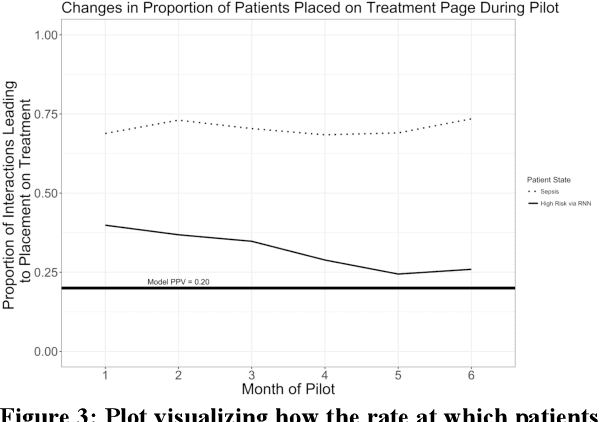
Abstract:Machine learning technologies are increasingly developed for use in healthcare. While research communities have focused on creating state-of-the-art models, there has been less focus on real world implementation and the associated challenges to accuracy, fairness, accountability, and transparency that come from actual, situated use. Serious questions remain under examined regarding how to ethically build models, interpret and explain model output, recognize and account for biases, and minimize disruptions to professional expertise and work cultures. We address this gap in the literature and provide a detailed case study covering the development, implementation, and evaluation of Sepsis Watch, a machine learning-driven tool that assists hospital clinicians in the early diagnosis and treatment of sepsis. We, the team that developed and evaluated the tool, discuss our conceptualization of the tool not as a model deployed in the world but instead as a socio-technical system requiring integration into existing social and professional contexts. Rather than focusing on model interpretability to ensure a fair and accountable machine learning, we point toward four key values and practices that should be considered when developing machine learning to support clinical decision-making: rigorously define the problem in context, build relationships with stakeholders, respect professional discretion, and create ongoing feedback loops with stakeholders. Our work has significant implications for future research regarding mechanisms of institutional accountability and considerations for designing machine learning systems. Our work underscores the limits of model interpretability as a solution to ensure transparency, accuracy, and accountability in practice. Instead, our work demonstrates other means and goals to achieve FATML values in design and in practice.
An Improved Multi-Output Gaussian Process RNN with Real-Time Validation for Early Sepsis Detection
Aug 19, 2017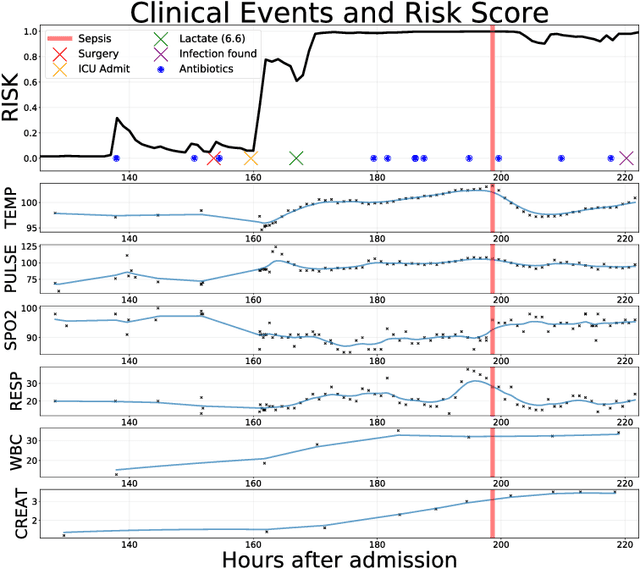
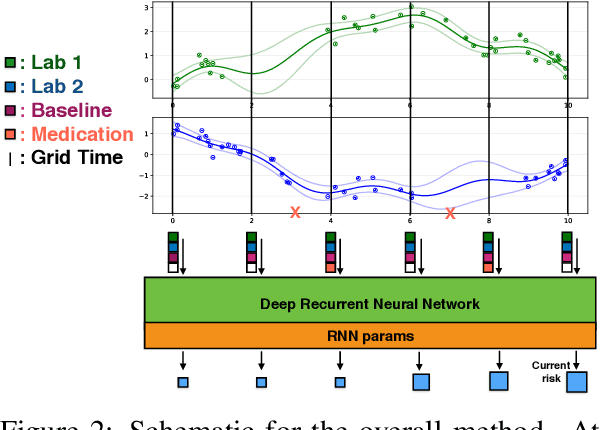
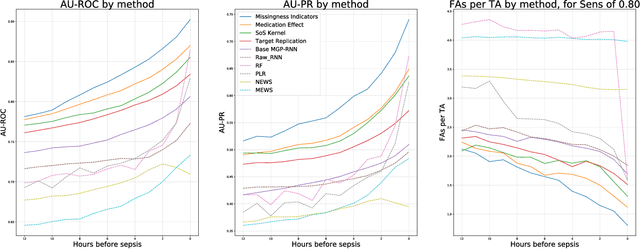
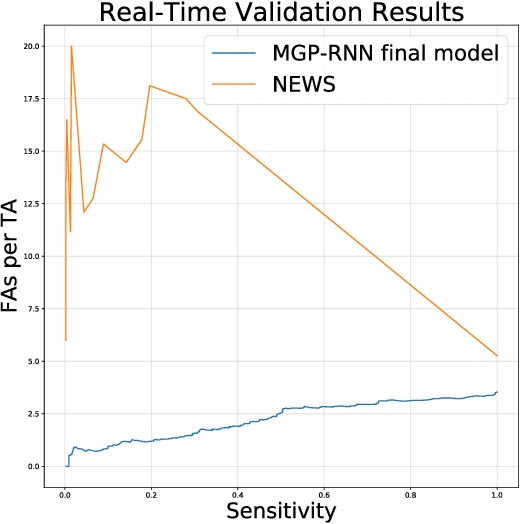
Abstract:Sepsis is a poorly understood and potentially life-threatening complication that can occur as a result of infection. Early detection and treatment improves patient outcomes, and as such it poses an important challenge in medicine. In this work, we develop a flexible classifier that leverages streaming lab results, vitals, and medications to predict sepsis before it occurs. We model patient clinical time series with multi-output Gaussian processes, maintaining uncertainty about the physiological state of a patient while also imputing missing values. The mean function takes into account the effects of medications administered on the trajectories of the physiological variables. Latent function values from the Gaussian process are then fed into a deep recurrent neural network to classify patient encounters as septic or not, and the overall model is trained end-to-end using back-propagation. We train and validate our model on a large dataset of 18 months of heterogeneous inpatient stays from the Duke University Health System, and develop a new "real-time" validation scheme for simulating the performance of our model as it will actually be used. Our proposed method substantially outperforms clinical baselines, and improves on a previous related model for detecting sepsis. Our model's predictions will be displayed in a real-time analytics dashboard to be used by a sepsis rapid response team to help detect and improve treatment of sepsis.
Learning to Detect Sepsis with a Multitask Gaussian Process RNN Classifier
Jun 13, 2017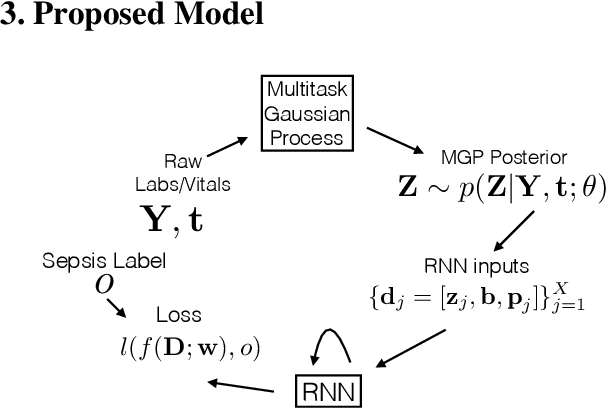
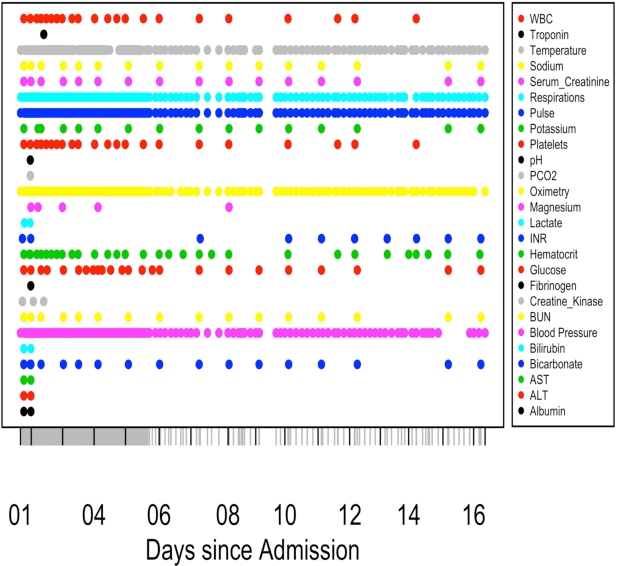
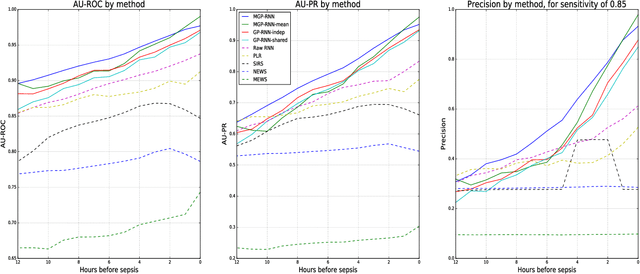
Abstract:We present a scalable end-to-end classifier that uses streaming physiological and medication data to accurately predict the onset of sepsis, a life-threatening complication from infections that has high mortality and morbidity. Our proposed framework models the multivariate trajectories of continuous-valued physiological time series using multitask Gaussian processes, seamlessly accounting for the high uncertainty, frequent missingness, and irregular sampling rates typically associated with real clinical data. The Gaussian process is directly connected to a black-box classifier that predicts whether a patient will become septic, chosen in our case to be a recurrent neural network to account for the extreme variability in the length of patient encounters. We show how to scale the computations associated with the Gaussian process in a manner so that the entire system can be discriminatively trained end-to-end using backpropagation. In a large cohort of heterogeneous inpatient encounters at our university health system we find that it outperforms several baselines at predicting sepsis, and yields 19.4% and 55.5% improved areas under the Receiver Operating Characteristic and Precision Recall curves as compared to the NEWS score currently used by our hospital.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge