Armando Bedoya
"The Human Body is a Black Box": Supporting Clinical Decision-Making with Deep Learning
Dec 07, 2019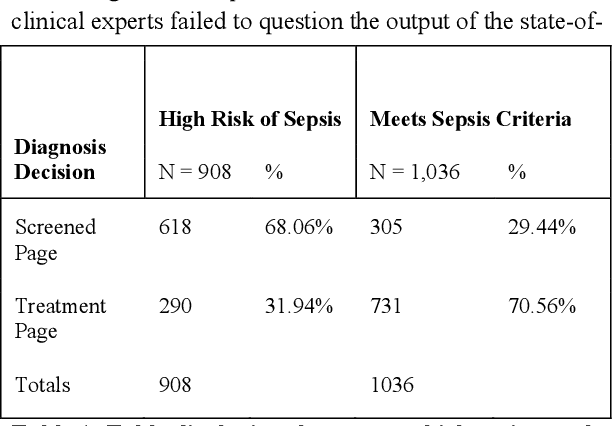
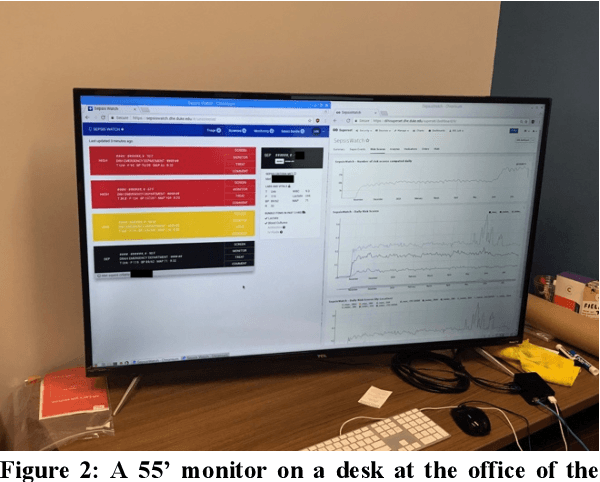
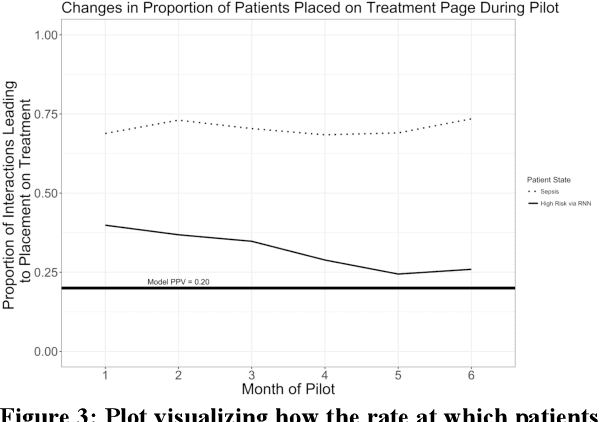
Abstract:Machine learning technologies are increasingly developed for use in healthcare. While research communities have focused on creating state-of-the-art models, there has been less focus on real world implementation and the associated challenges to accuracy, fairness, accountability, and transparency that come from actual, situated use. Serious questions remain under examined regarding how to ethically build models, interpret and explain model output, recognize and account for biases, and minimize disruptions to professional expertise and work cultures. We address this gap in the literature and provide a detailed case study covering the development, implementation, and evaluation of Sepsis Watch, a machine learning-driven tool that assists hospital clinicians in the early diagnosis and treatment of sepsis. We, the team that developed and evaluated the tool, discuss our conceptualization of the tool not as a model deployed in the world but instead as a socio-technical system requiring integration into existing social and professional contexts. Rather than focusing on model interpretability to ensure a fair and accountable machine learning, we point toward four key values and practices that should be considered when developing machine learning to support clinical decision-making: rigorously define the problem in context, build relationships with stakeholders, respect professional discretion, and create ongoing feedback loops with stakeholders. Our work has significant implications for future research regarding mechanisms of institutional accountability and considerations for designing machine learning systems. Our work underscores the limits of model interpretability as a solution to ensure transparency, accuracy, and accountability in practice. Instead, our work demonstrates other means and goals to achieve FATML values in design and in practice.
An Improved Multi-Output Gaussian Process RNN with Real-Time Validation for Early Sepsis Detection
Aug 19, 2017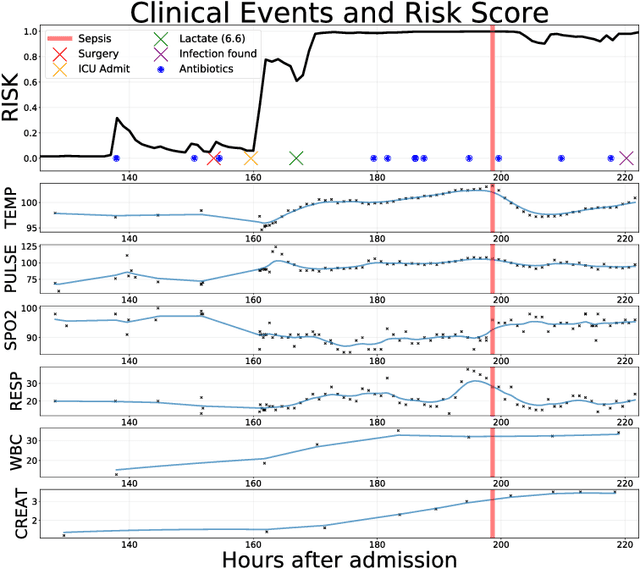
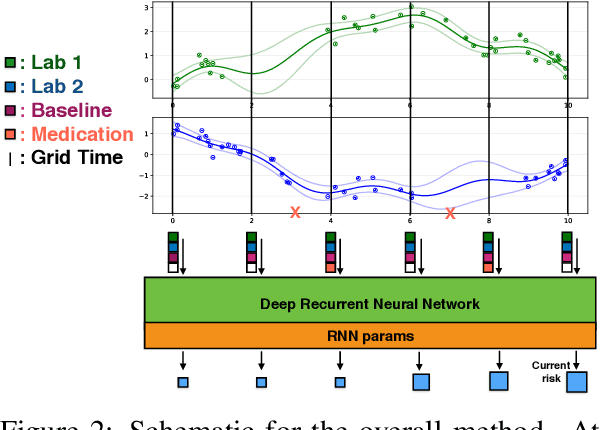
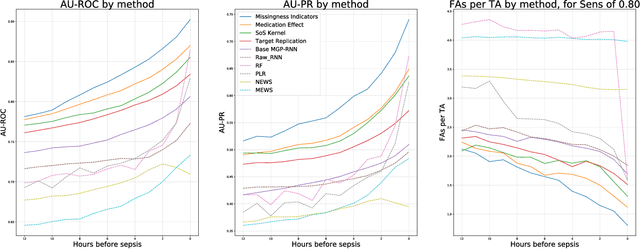
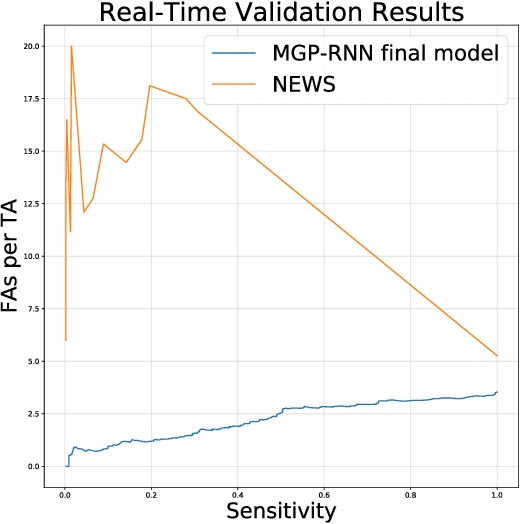
Abstract:Sepsis is a poorly understood and potentially life-threatening complication that can occur as a result of infection. Early detection and treatment improves patient outcomes, and as such it poses an important challenge in medicine. In this work, we develop a flexible classifier that leverages streaming lab results, vitals, and medications to predict sepsis before it occurs. We model patient clinical time series with multi-output Gaussian processes, maintaining uncertainty about the physiological state of a patient while also imputing missing values. The mean function takes into account the effects of medications administered on the trajectories of the physiological variables. Latent function values from the Gaussian process are then fed into a deep recurrent neural network to classify patient encounters as septic or not, and the overall model is trained end-to-end using back-propagation. We train and validate our model on a large dataset of 18 months of heterogeneous inpatient stays from the Duke University Health System, and develop a new "real-time" validation scheme for simulating the performance of our model as it will actually be used. Our proposed method substantially outperforms clinical baselines, and improves on a previous related model for detecting sepsis. Our model's predictions will be displayed in a real-time analytics dashboard to be used by a sepsis rapid response team to help detect and improve treatment of sepsis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge